Communicating a data analysis
1 / 26
by Dr. Lucy D'Agostino McGowan
Agenda
- Updating your
yaml - Putting all code in the appendix
- Hiding messages and warnings from your package loadings
- Hiding interactive output
- Creating pretty tables
- Creating figure and table caption
- Putting intermediate figures at the end
- Including citations
2 / 26
by Dr. Lucy D'Agostino McGowan
Updating your yaml
- For the final project I want you to create .pdf files.
---title: "The title of your document"name: "Your name"output: pdf_document---3 / 26
by Dr. Lucy D'Agostino McGowan
Updating your yaml
- For the final project I want you to create .pdf files.
---title: "The title of your document"name: "Your name"output: pdf_document fontsize: 12pt---- 12 pt font
4 / 26
by Dr. Lucy D'Agostino McGowan
Updating your yaml
- For the final project I want you to create .pdf files.
---title: "The title of your document"name: "Your name"output: pdf_document fontsize: 12ptlinestretch: 2---- 12 pt font
- Double spaced
5 / 26
by Dr. Lucy D'Agostino McGowan
Putting all code in the Appendix
- For these fancy reports, we want to see the R code, but we don't want it intersperced throughout the document.
- You can "hide" all of your chunks by adding this to the top of your .Rmd file
```{r, echo = FALSE}library(knitr)opts_chunk$set(echo = FALSE)```6 / 26
by Dr. Lucy D'Agostino McGowan
Putting all code in the Appendix
- For these fancy reports, we want to see the R code, but we don't want it intersperced throughout the document.
- You can "hide" all of your chunks by adding this to the top of your .Rmd file
```{r, echo = FALSE}library(knitr)opts_chunk$set(echo = FALSE)```- THEN at the end of you document (in the Appendix) you can add
```{r ref.label=knitr::all_labels(), echo = TRUE, eval = FALSE}```6 / 26
by Dr. Lucy D'Agostino McGowan
Hiding messages and warnings from your package loadings
- You can hide any messages or warnings by updating the chunk options
```{r, echo = FALSE}library(knitr)opts_chunk$set(echo = FALSE, message = FALSE, warning = FALSE)```7 / 26
by Dr. Lucy D'Agostino McGowan
Hiding interactive output
- If you are running "interactive code", that is code that is not saved to an object in
R, it will print in your output, even if you hide the code withecho = FALSE. For example:
glimpse(iris)## Observations: 150## Variables: 5## $ Sepal.Length <dbl> 5.1, 4.9, 4.7, 4.6, 5.0, 5.4, 4.6, 5.0, 4.4, 4.9, 5…## $ Sepal.Width <dbl> 3.5, 3.0, 3.2, 3.1, 3.6, 3.9, 3.4, 3.4, 2.9, 3.1, 3…## $ Petal.Length <dbl> 1.4, 1.4, 1.3, 1.5, 1.4, 1.7, 1.4, 1.5, 1.4, 1.5, 1…## $ Petal.Width <dbl> 0.2, 0.2, 0.2, 0.2, 0.2, 0.4, 0.3, 0.2, 0.2, 0.1, 0…## $ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, set…8 / 26
by Dr. Lucy D'Agostino McGowan
Hiding interactive output
- If you are running "interactive code", that is code that is not saved to an object in
R, it will print in your output, even if you hide the code withecho = FALSE. For example:
glimpse(iris)## Observations: 150## Variables: 5## $ Sepal.Length <dbl> 5.1, 4.9, 4.7, 4.6, 5.0, 5.4, 4.6, 5.0, 4.4, 4.9, 5…## $ Sepal.Width <dbl> 3.5, 3.0, 3.2, 3.1, 3.6, 3.9, 3.4, 3.4, 2.9, 3.1, 3…## $ Petal.Length <dbl> 1.4, 1.4, 1.3, 1.5, 1.4, 1.7, 1.4, 1.5, 1.4, 1.5, 1…## $ Petal.Width <dbl> 0.2, 0.2, 0.2, 0.2, 0.2, 0.4, 0.3, 0.2, 0.2, 0.1, 0…## $ Species <fct> setosa, setosa, setosa, setosa, setosa, setosa, set…- Use the
results = "hide"in the chunk options to hide it.
```{r, results = "hide"}glimpse(iris)```8 / 26
by Dr. Lucy D'Agostino McGowan
Creating pretty tables
- We've been using the
tidy()andglance()functions to print tables of our model output. - These kind of look like "code" in our final reports
- We can make these prettier with the knitr package
9 / 26
by Dr. Lucy D'Agostino McGowan
Creating pretty tables
- We've been using the
tidy()andglance()functions to print tables of our model output. - These kind of look like "code" in our final reports
- We can make these prettier with the knitr package
- Load the knitr package with
library(knitr) - Use the
kable()function to output a pretty table
library(knitr)lm(wt ~ am, data = mtcars) %>% tidy() %>% kable()| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 3.77 | 0.165 | 22.89 | 0 |
| am | -1.36 | 0.258 | -5.26 | 0 |
9 / 26
by Dr. Lucy D'Agostino McGowan
Creating table captions
library(knitr) lm(wt ~ am, data = mtcars) %>% tidy() %>% kable(caption = "Predicting Car's weight from transmission type")| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | 3.77 | 0.165 | 22.89 | 0 |
| am | -1.36 | 0.258 | -5.26 | 0 |
10 / 26
by Dr. Lucy D'Agostino McGowan
Creating figure captions
- If you have an
Rchunck that produces a Figure, you can caption it using thefig.capchunk option. For example:
ggplot(mtcars, aes(x = mpg, y = disp)) + geom_point()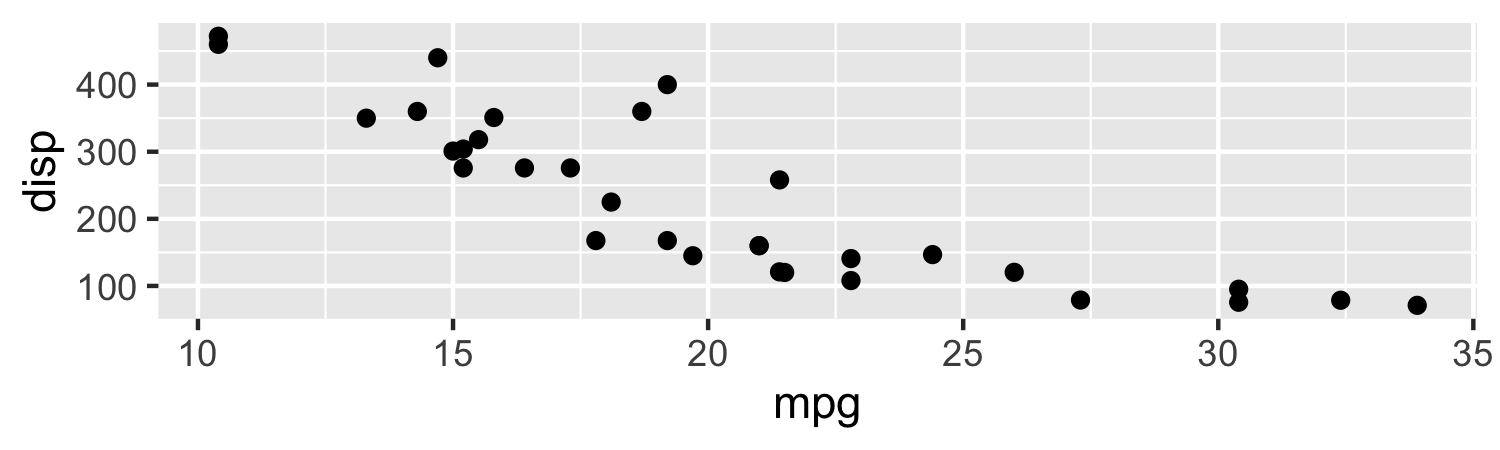
```{r, fig.cap = "Scatterplot of Miles per gallon by Displacement"}ggplot(mtcars, aes(x = mpg, y = disp)) + geom_point()```11 / 26
by Dr. Lucy D'Agostino McGowan
Putting extra figures at the end of your document
- Your prompt tells you to put all "intermediate" figures in the appendix
- This includes figures that you used to check the assumptions of the models that were not the final model
- To do this:
- Save your figures as
Robjects - Create code chunks at the beginning of the Appendix that run these objects (with captions)
- Save your figures as
12 / 26
by Dr. Lucy D'Agostino McGowan
Putting extra figures at the end of your document
- Save the plot to an
Robject,p
p <- ggplot(mtcars, aes(mpg, disp)) + geom_point()13 / 26
by Dr. Lucy D'Agostino McGowan
Putting extra figures at the end of your document
- Save the plot to an
Robject,p
p <- ggplot(mtcars, aes(mpg, disp)) + geom_point()- Put this
Robject,pin a chunk at the beginning of your Appendix
```{r, fig.cap = "Scatterplot of Miles per gallon by Displacement"}p```13 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- Your introduction should cite prior research in the area of your research question
- You will need to create a
citations.bibfile with citations, and then reference them using[@citation]
14 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- Create a
citations.bibfile in your RStudio project- Go to File > New File > Text File and click
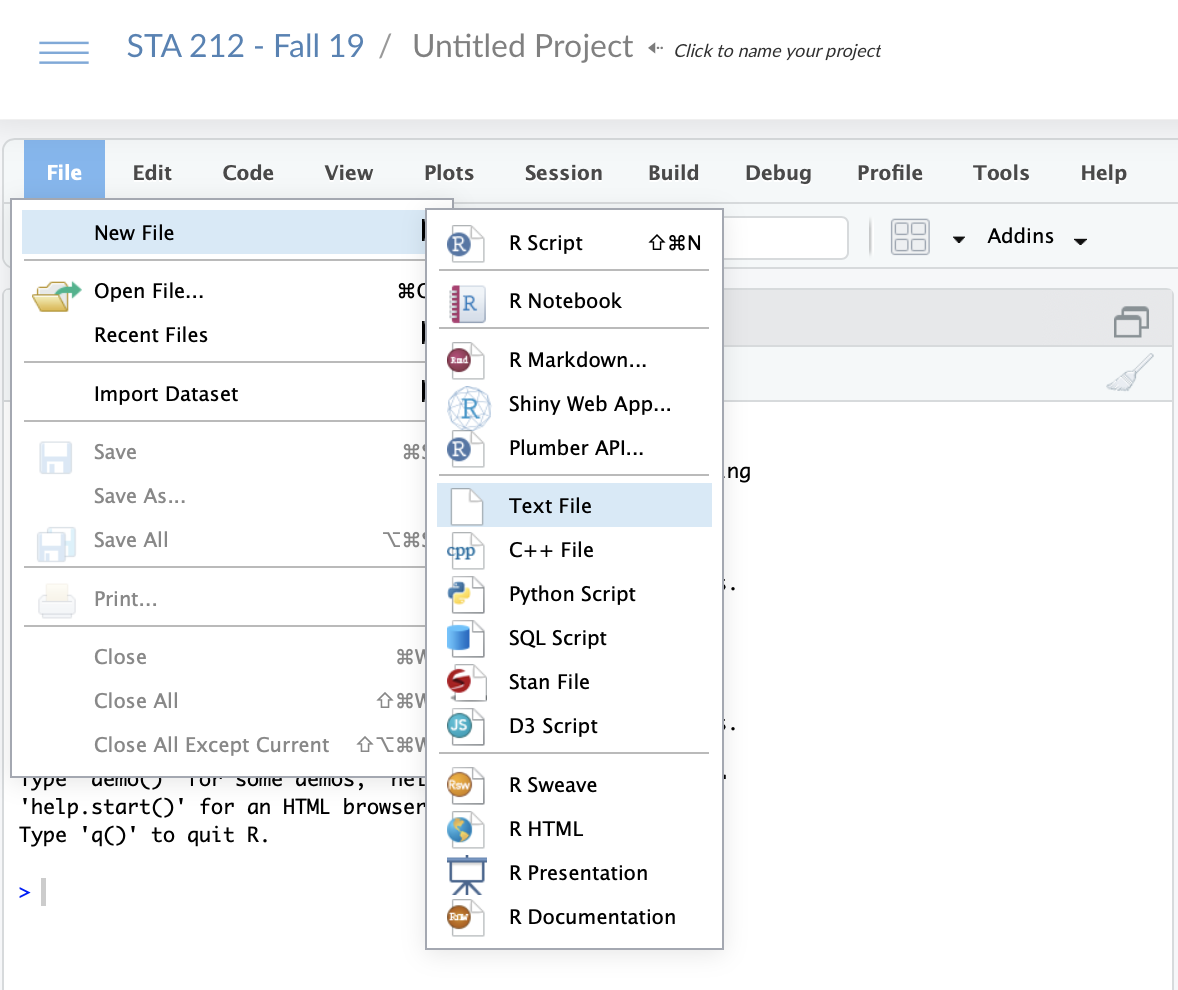
15 / 26
by Dr. Lucy D'Agostino McGowan
Citations
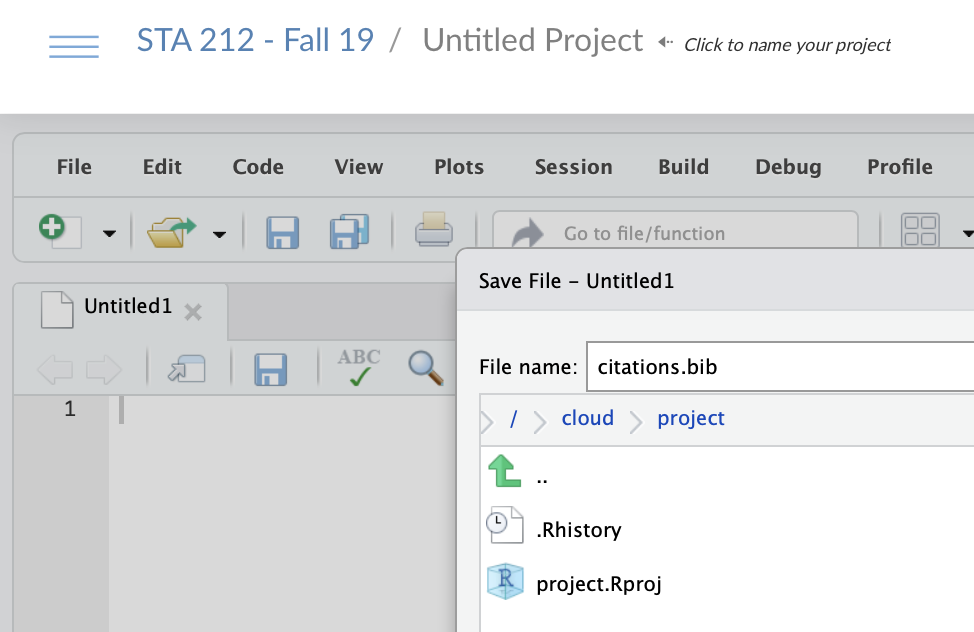
- Create a
citations.bibfile in your RStudio project- Go to File > New File > Text File and click
- Save the file as
citations.bib
16 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- This bibliography is using
BibTex, which is a certain way citations are saved in Latex - Your
citations.bibis going to be a file with multipleBibTexentries - You can get
Bibtexentries from Google Scholar- Search for the article you'd like to cite
- Under the article there will be
", click that

17 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- This bibliography is using
BibTex, which is a certain way citations are saved in Latex - Your
citations.bibis going to be a file with multipleBibTexentries - You can get
Bibtexentries from Google Scholar- Search for the article you'd like to cite
- Under the article there will be
", click that - A window will pop up with several citation styles, click
BibTexon the bottom
18 / 26
by Dr. Lucy D'Agostino McGowan
Citations

19 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- This bibliography is using
BibTex, which is a certain way citations are saved in Latex - Your
citations.bibis going to be a file with multipleBibTexentries - You can get
Bibtexentries from Google Scholar- Search for the article you'd like to cite
- Under the article there will be
", click that - A window will pop up with several citation styles, click
BibTexon the bottom - Copy the text in the window that pops up into your
citations.bibfile in RStudio
20 / 26
by Dr. Lucy D'Agostino McGowan
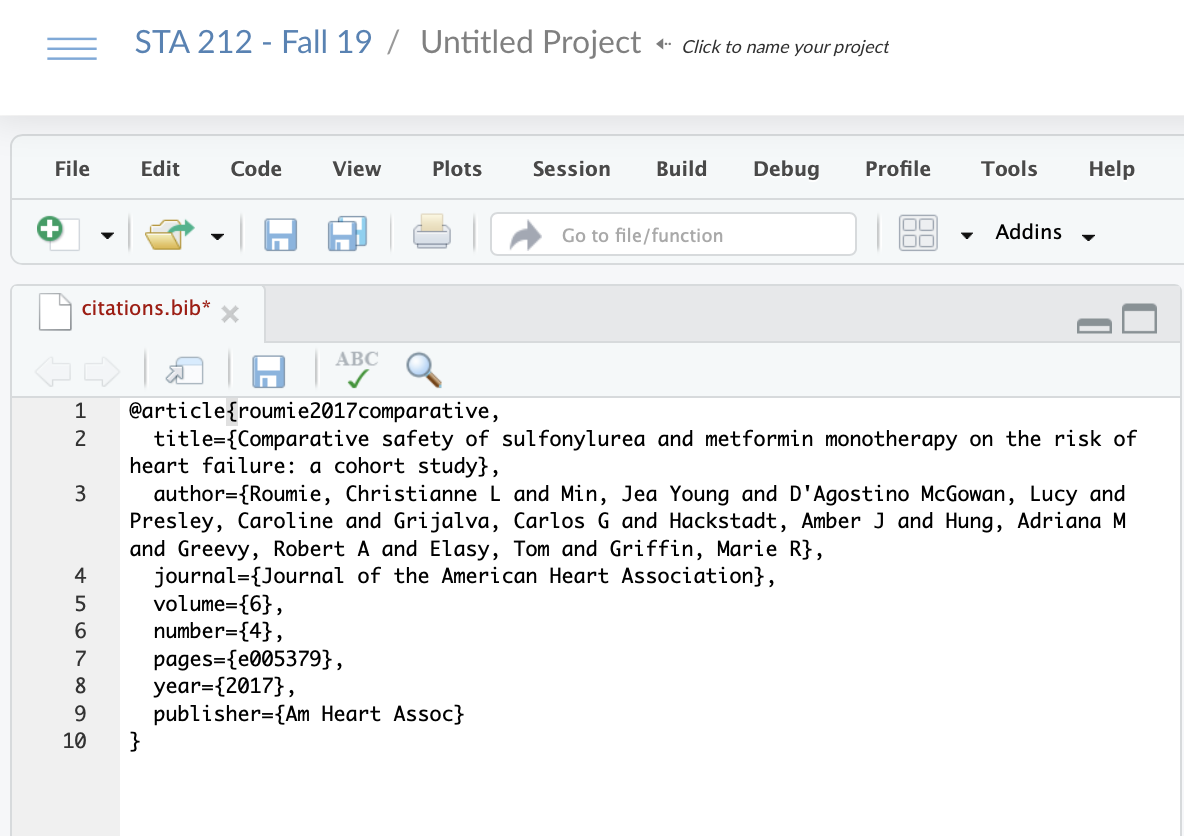
Citations
21 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- The first argument of this citation is the reference key that you will use in your main document. Here it is
roumie2017comparative
@article{roumie2017comparative, title={Comparative safety of sulfonylurea and metformin monotherapy on the risk of heart failure: a cohort study}, author={Roumie, Christianne L and Min, Jea Young and D'Agostino McGowan, Lucy and Presley, Caroline and Grijalva, Carlos G and Hackstadt, Amber J and Hung, Adriana M and Greevy, Robert A and Elasy, Tom and Griffin, Marie R}, journal={Journal of the American Heart Association}, volume={6}, number={4}, pages={e005379}, year={2017}, publisher={Am Heart Assoc}}22 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- Go back to your final report document
- Add the
citations.bibfile to youryaml
---title: "The title of your document"name: "Your name"output: pdf_document fontsize: 12ptlinestretch: 2 bibliography: citations.bib---23 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- Go back to your final report document
- Add the
citations.bibfile to youryaml - When you want to cite this paper in the main text, use the key like this:
[@roumie2017comparative] - At the VERY end of your document (after the Appendix, after your code chunks) add this header:
## References24 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- What if the thing you want to cite isn't on Google Scholar?
- Generate a
BibTexobject here:
http://www.citationmachine.net/bibtex/cite-a-website
25 / 26
by Dr. Lucy D'Agostino McGowan
Citations
- The first line of your results should say "All analysis is completed in R" and then cite the R packages you used.
- Run this to get the citation:
print(citation("tidyverse"), bibtex = TRUE)Make sure there is a reference *key, if not add one.
.small[
@Manual{, title = {tidyverse: Easily Install and Load the 'Tidyverse'}, author = {Hadley Wickham}, year = {2017}, note = {R package version 1.2.1}, url = {https://CRAN.R-project.org/package=tidyverse}, }]
Citations
- The first line of your results should say
"All analysis is completed in R"
[@tidyverse]and then cite the R packages you used. - Run this to get the citation:print(citation("tidyverse"), bibtex = TRUE)
- Make sure there is a reference key, if not add one.
.small[
@Manual{tidyverse, title = {tidyverse: Easily Install and Load the 'Tidyverse'}, author = {Hadley Wickham}, year = {2017}, note = {R package version 1.2.1}, url = {https://CRAN.R-project.org/package=tidyverse}, }]
Citations
- The first line of your results should say
"All analysis is completed in R"
[@tidyverse; @broom]and then cite the R packages you used. - For more than one, seperate with a
; - Run this to get the citation:print(citation("broom"), bibtex = TRUE)
- Make sure there is a reference key, if not add one.
Rstudio Cloud
- Open your project for the final report
- Update the
yamlto include 12 pt font, double spacing, and a bibliography - Practice creating figure captions
- Practice adding figures to the Appendix
- Create a
citations.bibfile - Practice citing an article
26 / 26