Partitioning variability
Partitioning variability

Why?
- y−¯y=(^y−¯y)+(y−^y)
Why?
y−¯y=(^y−¯y)+(y−^y)
∑(y−¯y)2=∑(^y−¯y)2+∑(y−^y)2
Why?
y−¯y=(^y−¯y)+(y−^y)
∑(y−¯y)2=∑(^y−¯y)2+∑(y−^y)2
SSTotal = SSModel + SSE
Degrees of freedom
SSTotal: n−1
Degrees of freedom
SSTotal: n−1
SSE: n−2
Degrees of freedom
SSTotal: n−1
SSE: n−2
SSModel: n−1=1+(n−2) - so 1!
Mean Squares
- MSModel=SSModel1
Mean Squares
- MSModel=SSModel1
- MSE=SSEn−2
F=MSModelMSE
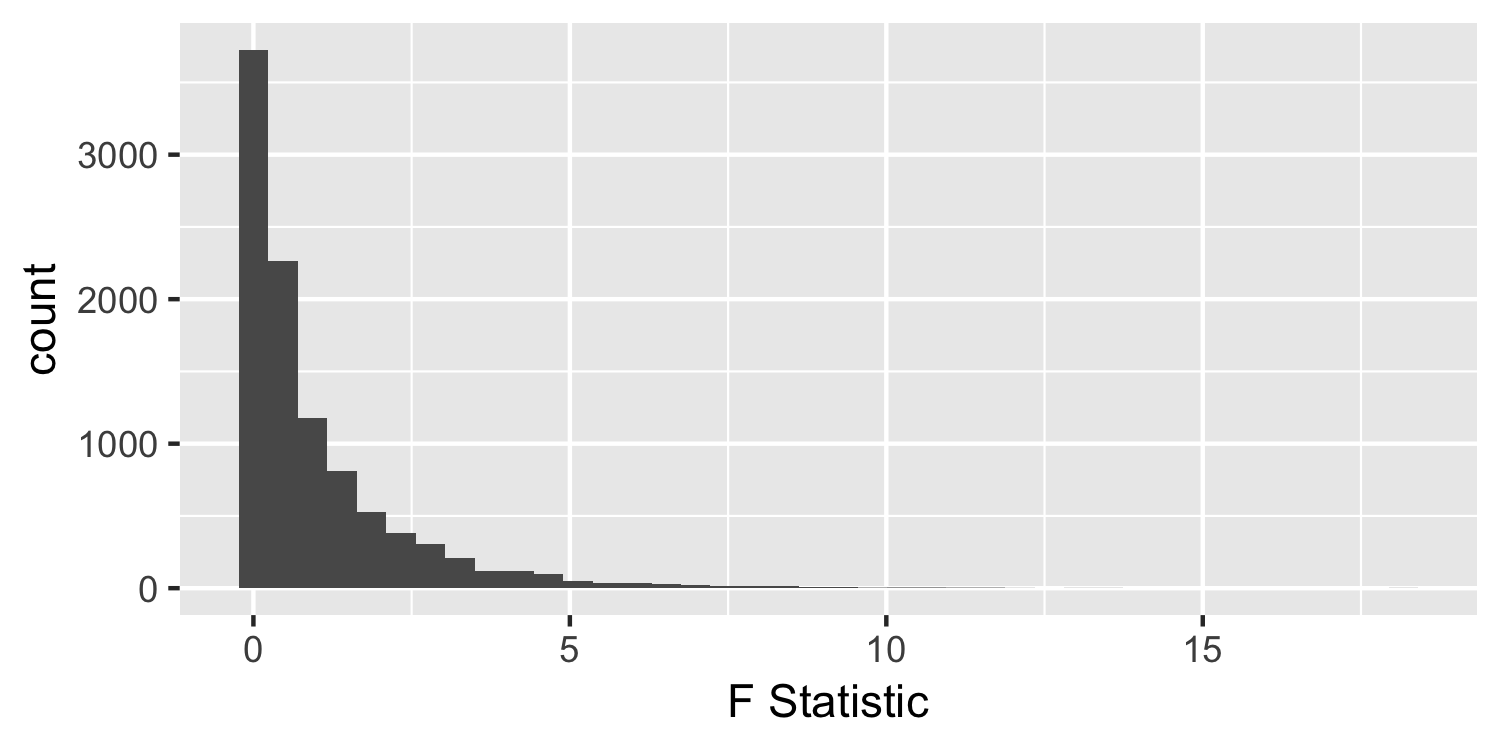
F-distribution
Under the null hypothesis
Sparrows
We can see all of these pieces using the anova() function
lm(Weight ~ WingLength, data = Sparrows) %>% anova()## Analysis of Variance Table## ## Response: Weight## Df Sum Sq Mean Sq F value Pr(>F)## WingLength 1 355.05 355.05 181.25 < 2.2e-16## Residuals 114 223.31 1.96Sparrows
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>Sparrows
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>- F-statistic: 181.25
Sparrows
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>- F-statistic: 181.25
- p-value: 2.62e-25
Sparrows
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>- F-statistic: 181.25
- p-value: 2.62e-25
- Where did this p-value come from?
p-value
The probability of getting a statistic as extreme or more extreme than the observed test statistic given the null hypothesis is true
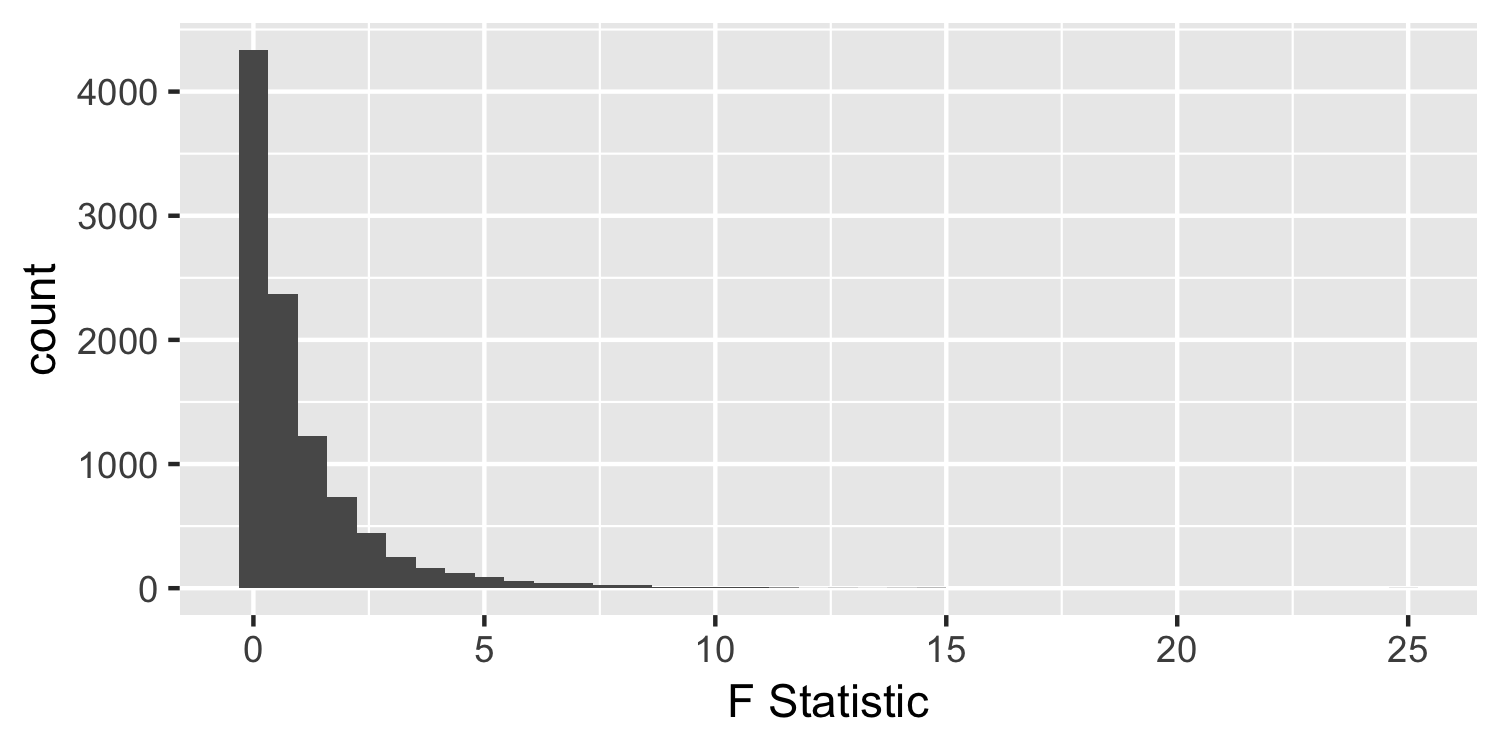
F-distribution
Under the null hypothesis
Sparrows: Degrees of freedom
- SSTotal: n−1 = 115
Sparrows: Degrees of freedom
- SSTotal: n−1 = 115
- SSE: ?
Sparrows: Degrees of freedom
- SSTotal: n−1 = 115
- SSE: ?
- SSModel: ?
Sparrows: Degrees of freedom
- SSTotal: n−1 = 115
Sparrows: Degrees of freedom
- SSTotal: n−1 = 115
- SSE: n−2 = 114
Sparrows: Degrees of freedom
- SSTotal: n−1 = 115
- SSE: n−2 = 114
- SSModel: 115 - 114 = 1
Sparrows
To calculate the p-value under the t-distribution we used pt(). What do you think we use to calculate the p-value under the F-distribution?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>Sparrows
To calculate the p-value under the t-distribution we used pt(). What do you think we use to calculate the p-value under the F-distribution?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>pf()
Sparrows
To calculate the p-value under the t-distribution we used pt(). What do you think we use to calculate the p-value under the F-distribution?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>pf()- it takes 3 arguments:
q,df1, anddf2. What do you thinkdf1anddf2are?
Sparrows
To calculate the p-value under the t-distribution we used pt(). What do you think we use to calculate the p-value under the F-distribution?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>pf(181.2535, 1, 114, lower.tail = FALSE)## [1] 2.621946e-25Sparrows
Why don't we multiple this p-value by 2 when we use pf()?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>pf(181.2535, 1, 114, lower.tail = FALSE)## [1] 2.621946e-25F-Distribution
Under the null hypothesis
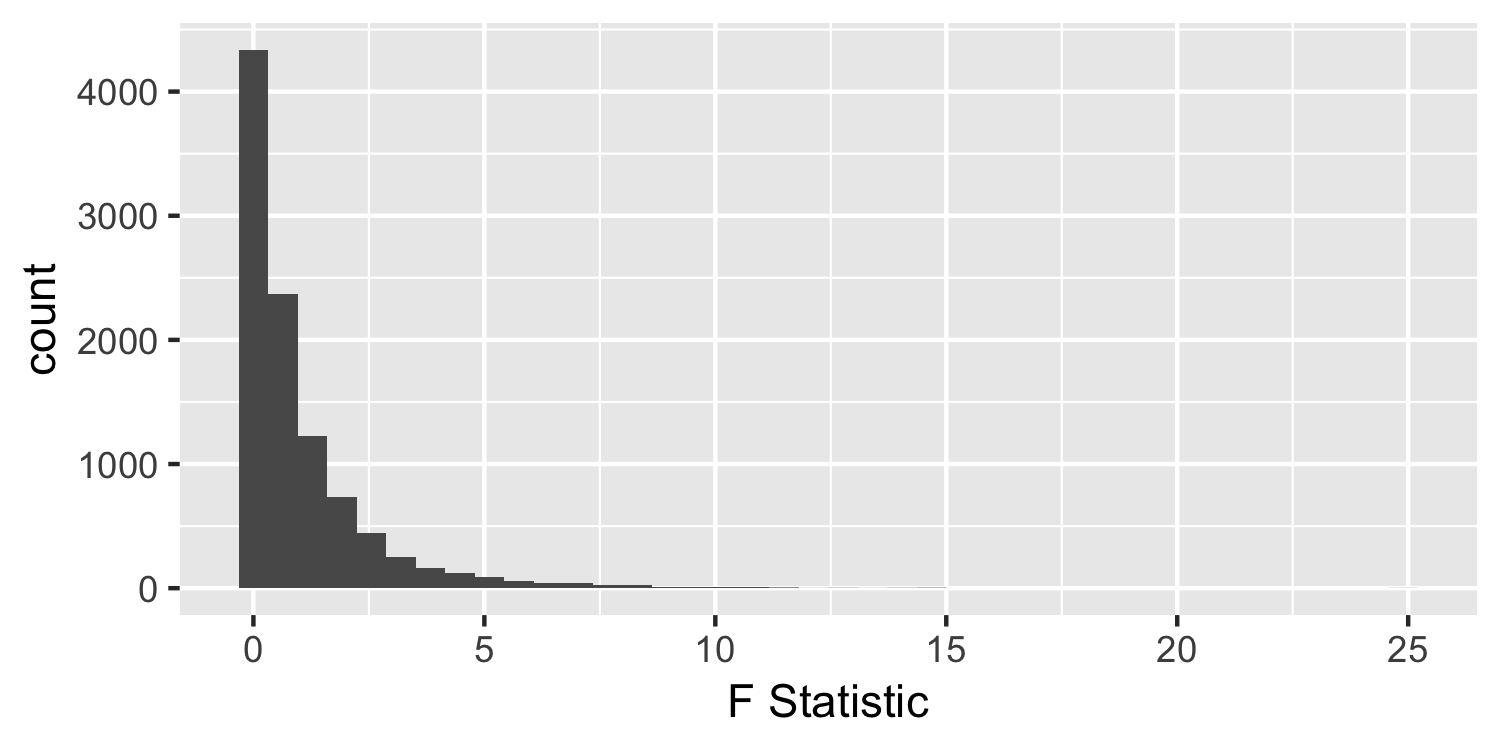
- We observed an F-statistic of 181.25, but for demonstration purposes, let's assume we saw one of 2.
F-Distribution
Under the null hypothesis
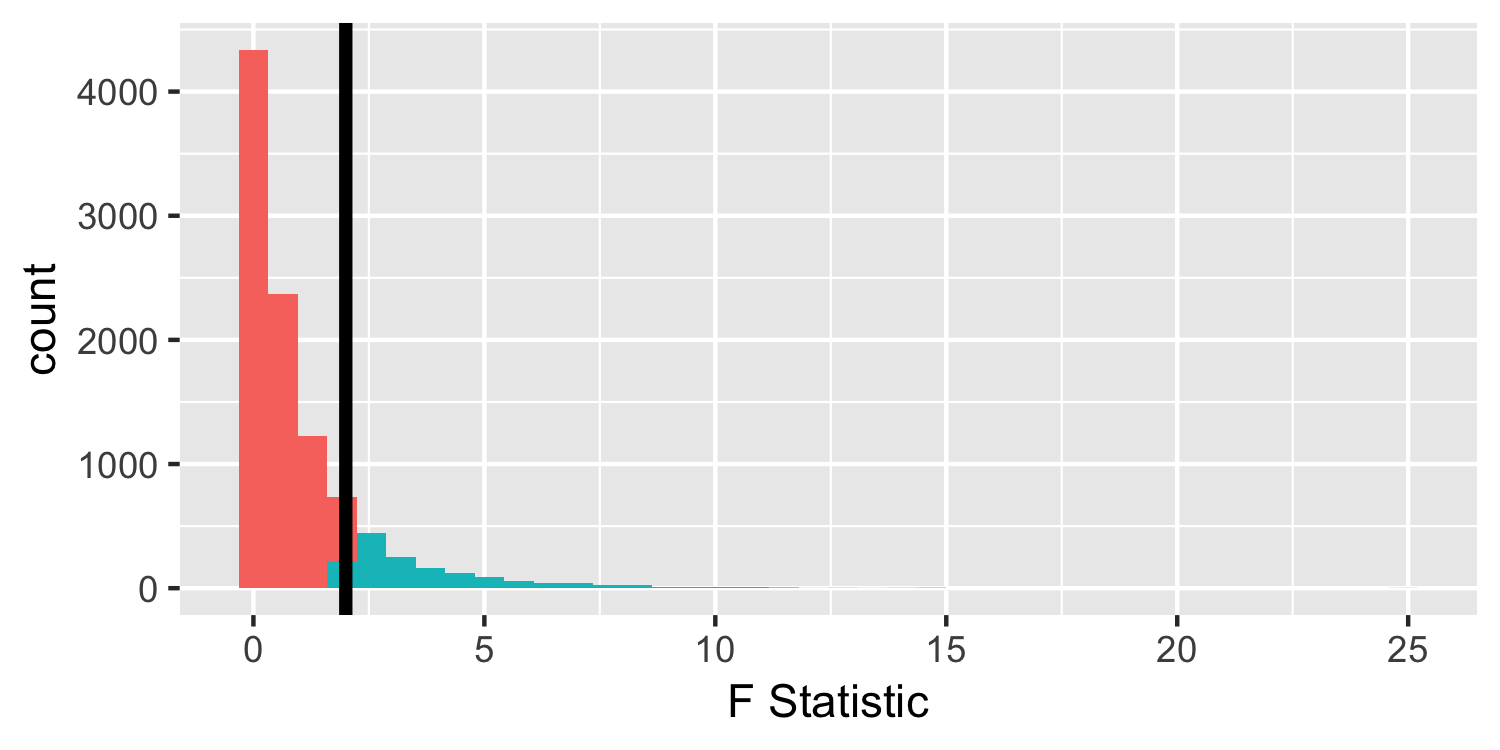
- We observed an F-statistic of 181.25, but for demonstration purposes, let's assume we saw one of 2.
F-Distribution
Under the null hypothesis
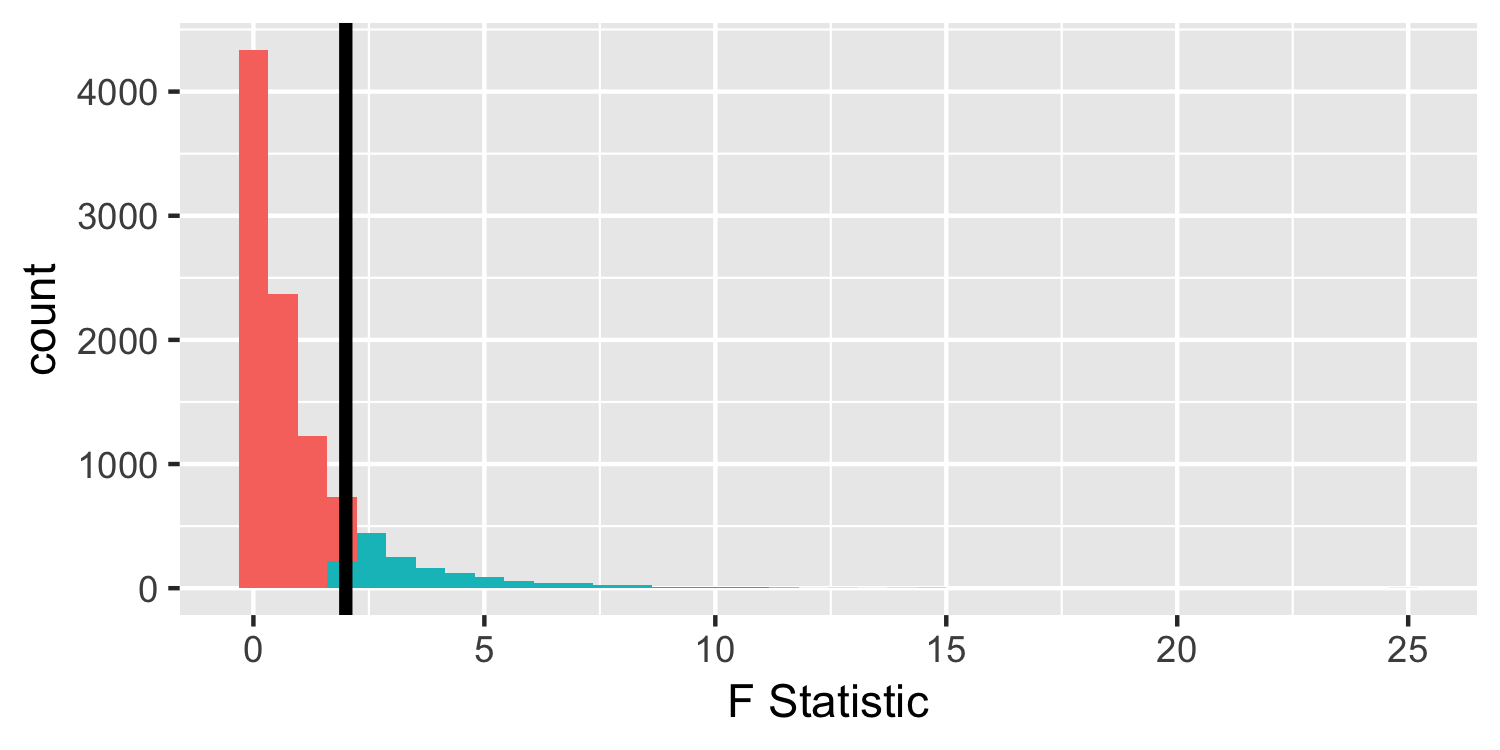
- Are there any negative values in an F-distribution?
F-Distribution
Under the null hypothesis
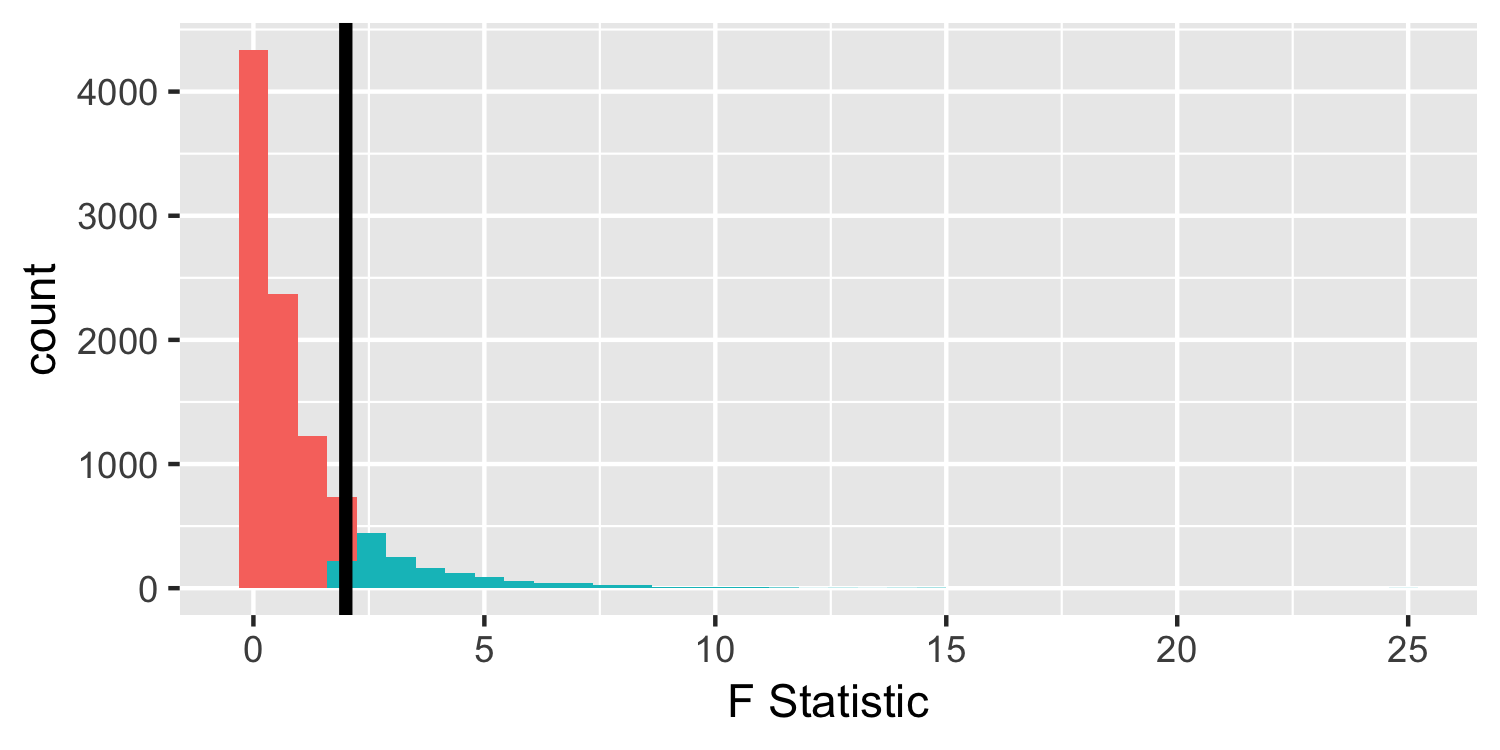
- The p-value calculates values "as extreme or more extreme", in the t-distribution "more extreme values", defined as farther from 0, can be positive or negative. Not so for the F!
Sparrows
Notice the p-value for the F-test is the same as the p-value for the t-test for β1!
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>lm(Weight ~ WingLength, data = Sparrows) %>% tidy()## # A tibble: 2 x 5## term estimate std.error statistic p.value## <chr> <dbl> <dbl> <dbl> <dbl>## 1 (Intercept) 1.37 0.957 1.43 1.56e- 1## 2 WingLength 0.467 0.0347 13.5 2.62e-25Sparrows
What is the F-test testing?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>Sparrows
What is the F-test testing?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>- null hypothesis: the fit of the intercept only model (with ^β0 only) and your model (in this case, ^β0+^β1x ) are equivalent
Sparrows
What is the F-test testing?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>- null hypothesis: the fit of the intercept only model (with ^β0 only) and your model (in this case, ^β0+^β1x ) are equivalent
- alternative hypothesis: The fit of the intercept only model is significantly worse compared to your model
Sparrows
What is the F-test testing?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>- null hypothesis: the fit of the intercept only model (with ^β0 only) and your model (in this case, ^β0+^β1x ) are equivalent
- alternative hypothesis: The fit of the intercept only model is significantly worse compared to your model
- When we only have one variable in our model, x, the p-values from the F and t are going to be equivalent
Sparrows
How are the test statistics related between the F and the t?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>lm(Weight ~ WingLength, data = Sparrows) %>% tidy()## # A tibble: 2 x 5## term estimate std.error statistic p.value## <chr> <dbl> <dbl> <dbl> <dbl>## 1 (Intercept) 1.37 0.957 1.43 1.56e- 1## 2 WingLength 0.467 0.0347 13.5 2.62e-25Sparrows
How are the test statistics related between the F and the t?
lm(Weight ~ WingLength, data = Sparrows) %>% glance()## # A tibble: 1 x 11## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC## <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>## 1 0.614 0.611 1.40 181. 2.62e-25 2 -203. 411. 419.## # … with 2 more variables: deviance <dbl>, df.residual <int>lm(Weight ~ WingLength, data = Sparrows) %>% tidy()## # A tibble: 2 x 5## term estimate std.error statistic p.value## <chr> <dbl> <dbl> <dbl> <dbl>## 1 (Intercept) 1.37 0.957 1.43 1.56e- 1## 2 WingLength 0.467 0.0347 13.5 2.62e-2513.5^2## [1] 182.25## [1] 182.25