Assessing Conditions
1 / 38
Dr. Lucy D'Agostino McGowan
Revisit late policy
2 / 38
Dr. Lucy D'Agostino McGowan
Porsche Price (2)
- Go to RStudio Cloud and open
Porsche Price (2)
3 / 38
Dr. Lucy D'Agostino McGowan
Steps for modeling
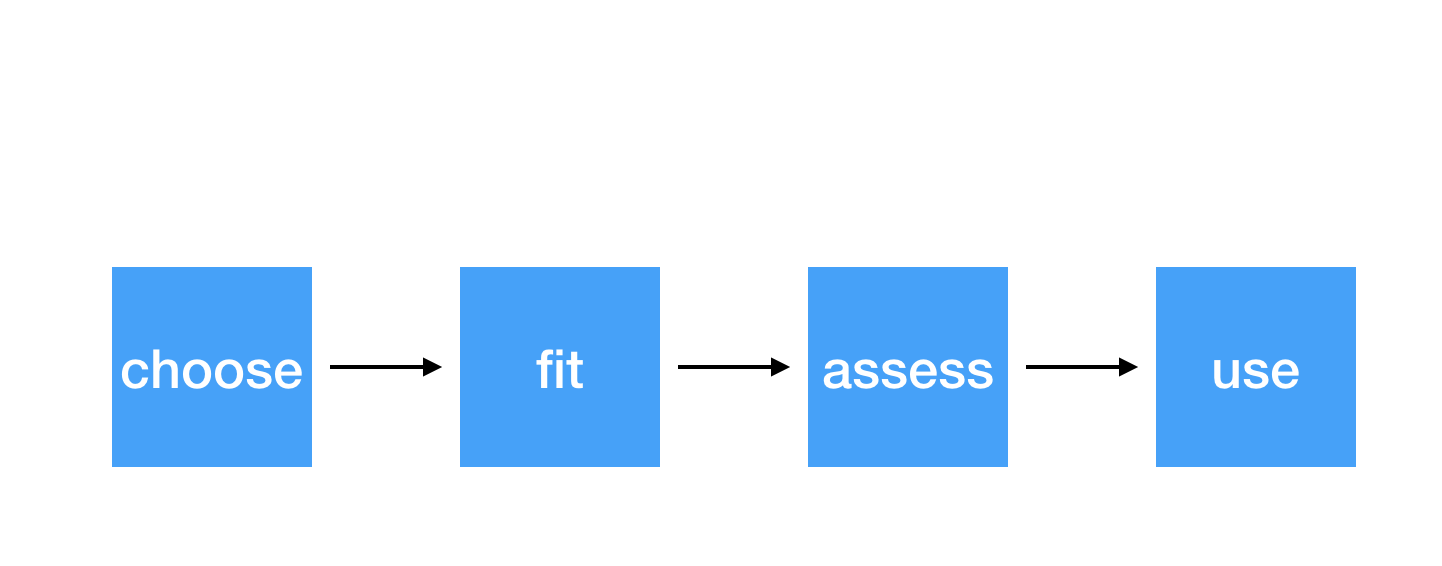
4 / 38
Dr. Lucy D'Agostino McGowan
Steps for modeling
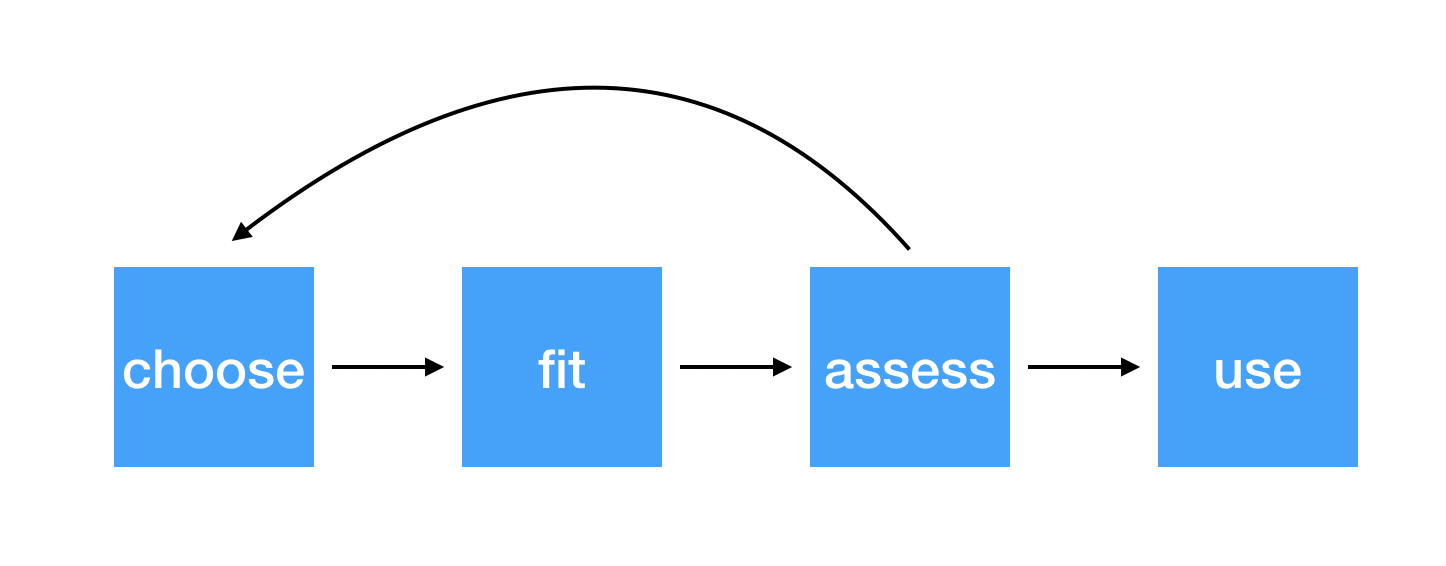
5 / 38
Dr. Lucy D'Agostino McGowan
Conditions for simple linear regression
- Linearity
- Zero Mean
- Constant Variance
- Independence
- Random
- Normality
6 / 38
Dr. Lucy D'Agostino McGowan
Conditions for simple linear regression
- Linearity
Zero Mean- Constant Variance
- Independence
- Random
- Normality
7 / 38
Dr. Lucy D'Agostino McGowan
Conditions for simple linear regression
- Linearity
Zero Mean- Constant Variance
- Independence
- Random
- Normality
8 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
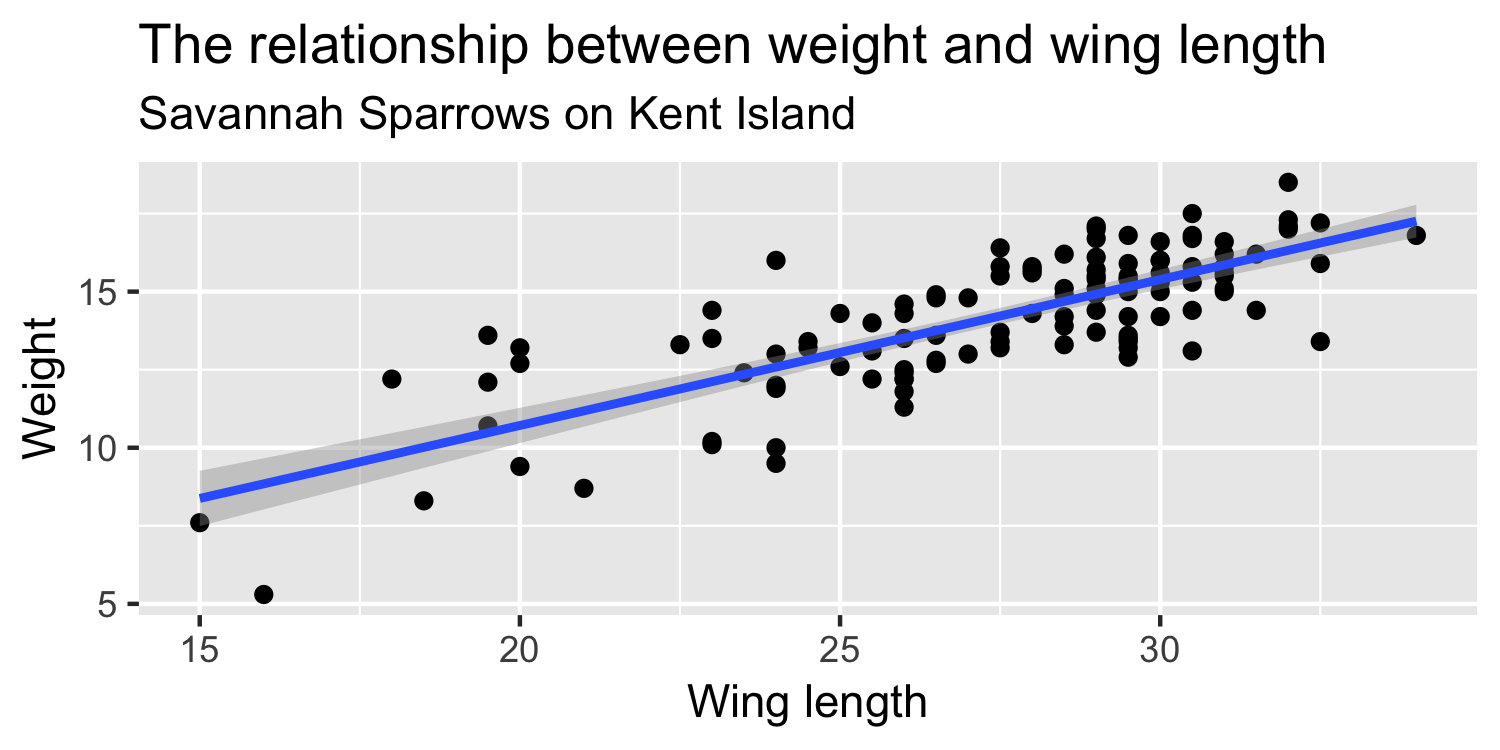
9 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
Residuals versus fits plot
What do you think would be on the x-axis and y-axis on a "residuals versus fits" plot?
10 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
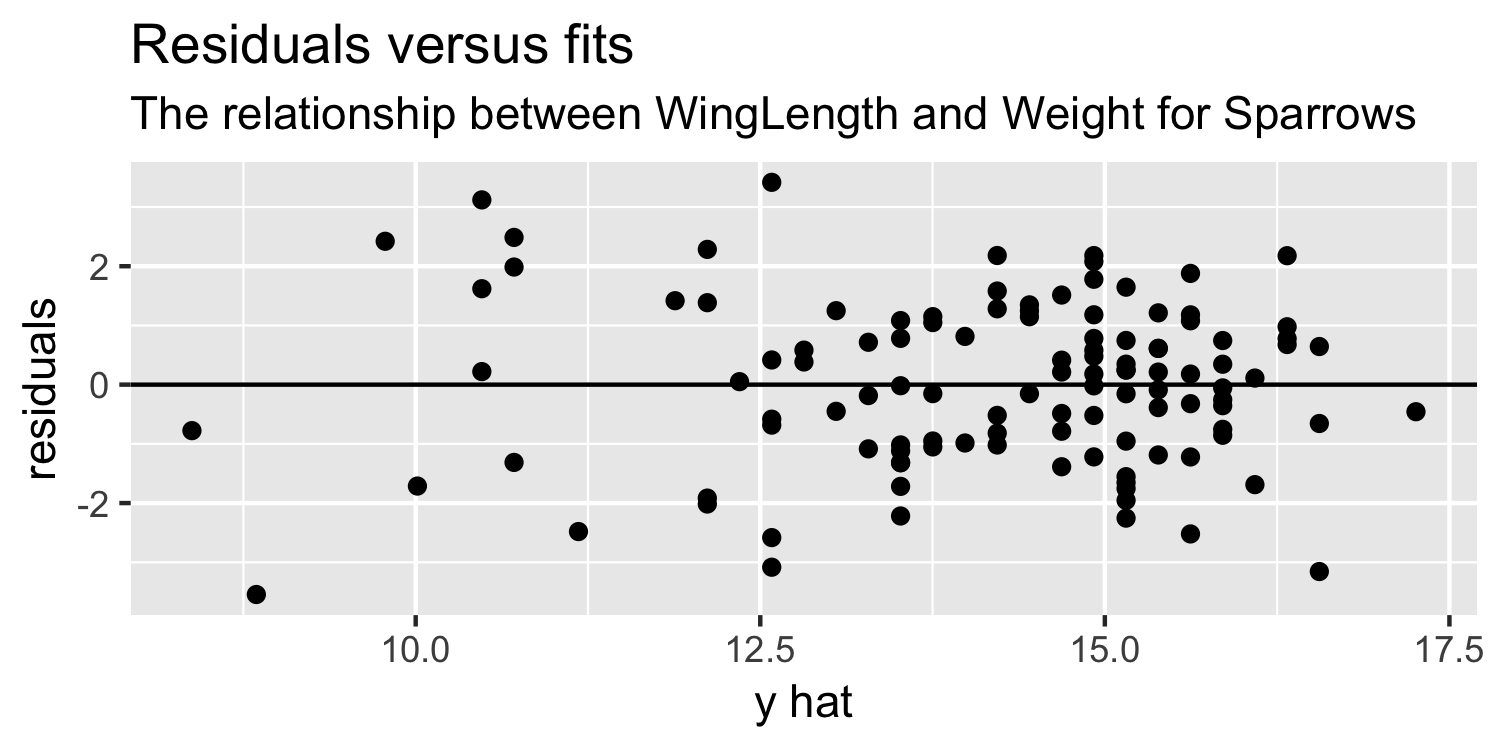
11 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
Residuals versus fits plot: What are we looking for?
- random variation above and below 0
- no apparent "patterns"
- the width of the "band" of points is relatively constant
12 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
What do you think of this plot?
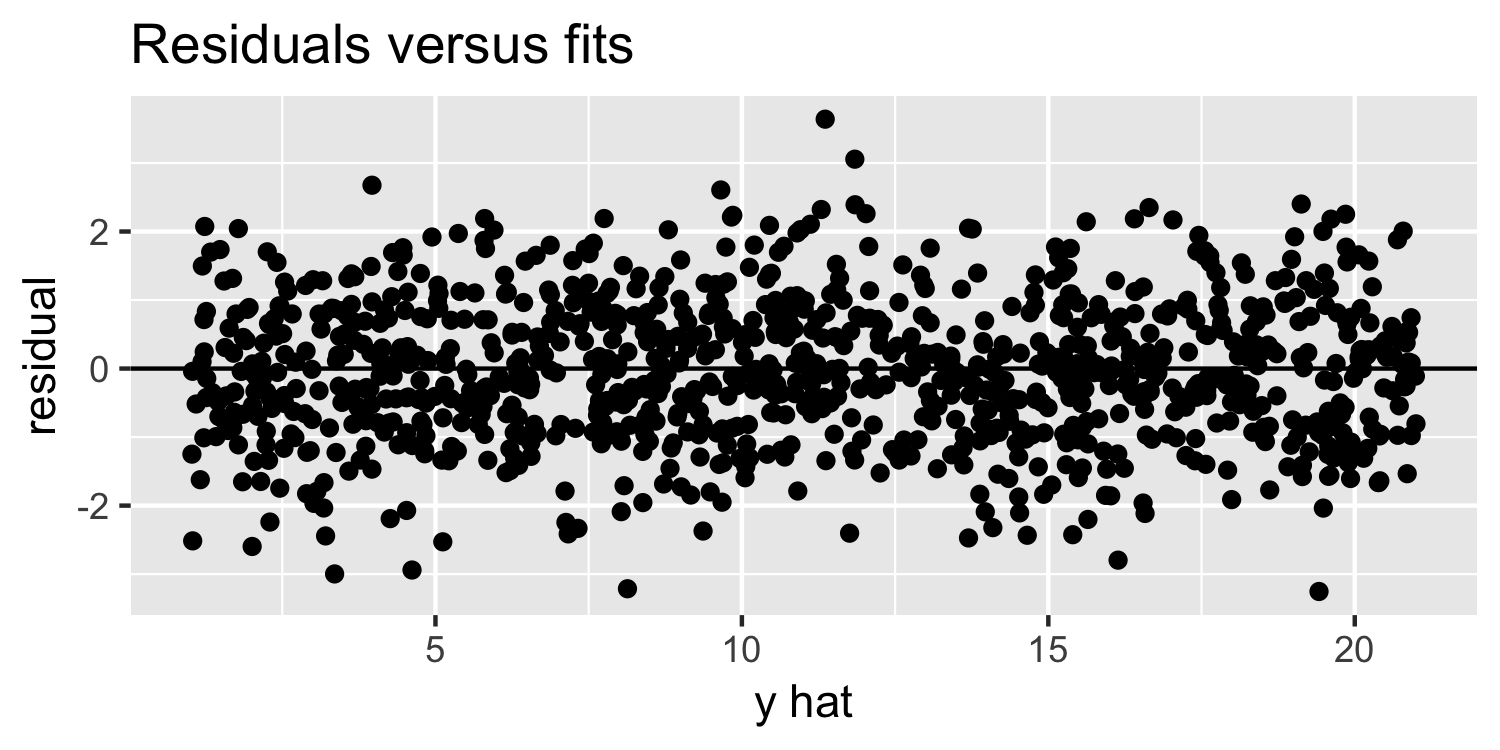
13 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
What do you think of this plot?
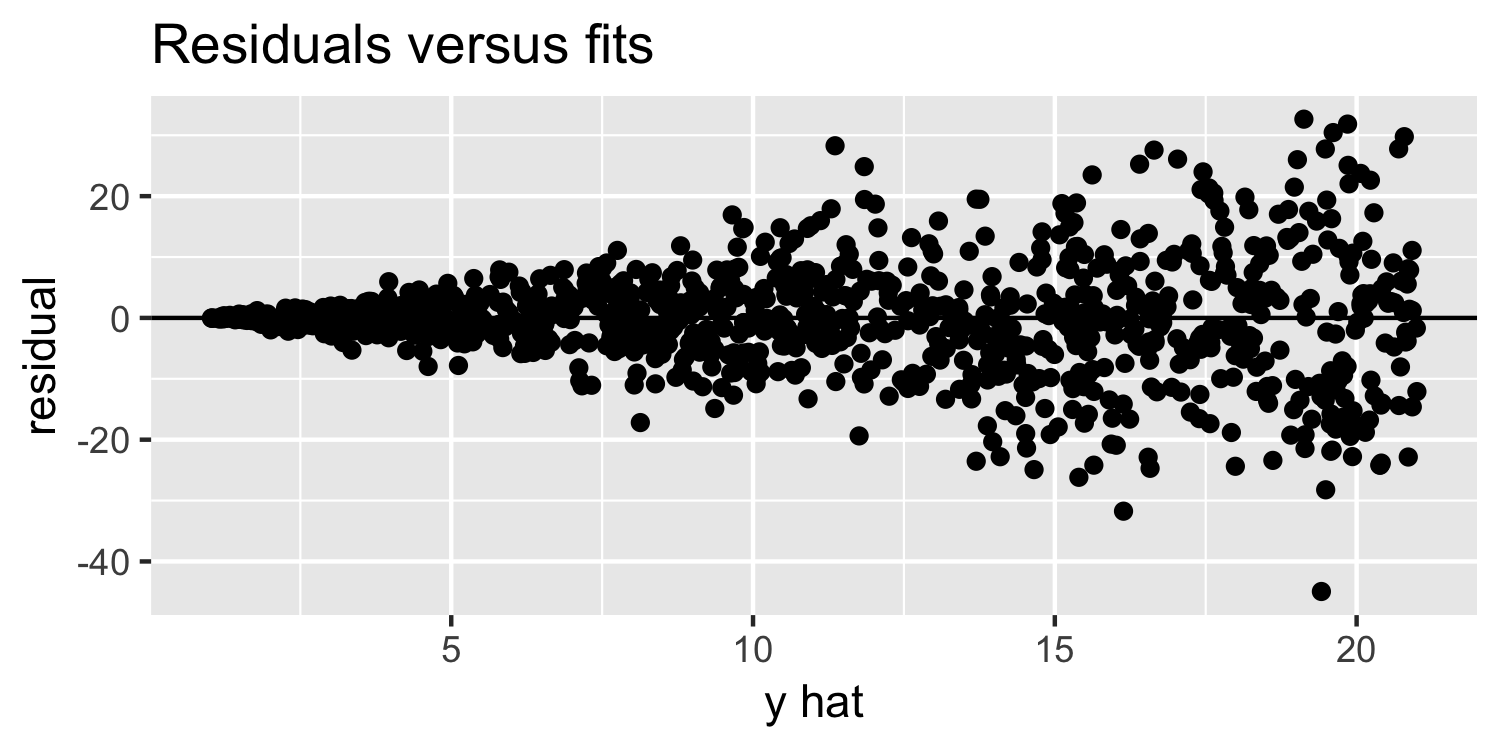
14 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
What do you think of this plot?

15 / 38
Dr. Lucy D'Agostino McGowan
Linearity & Constant Variance
What do you think of this plot?
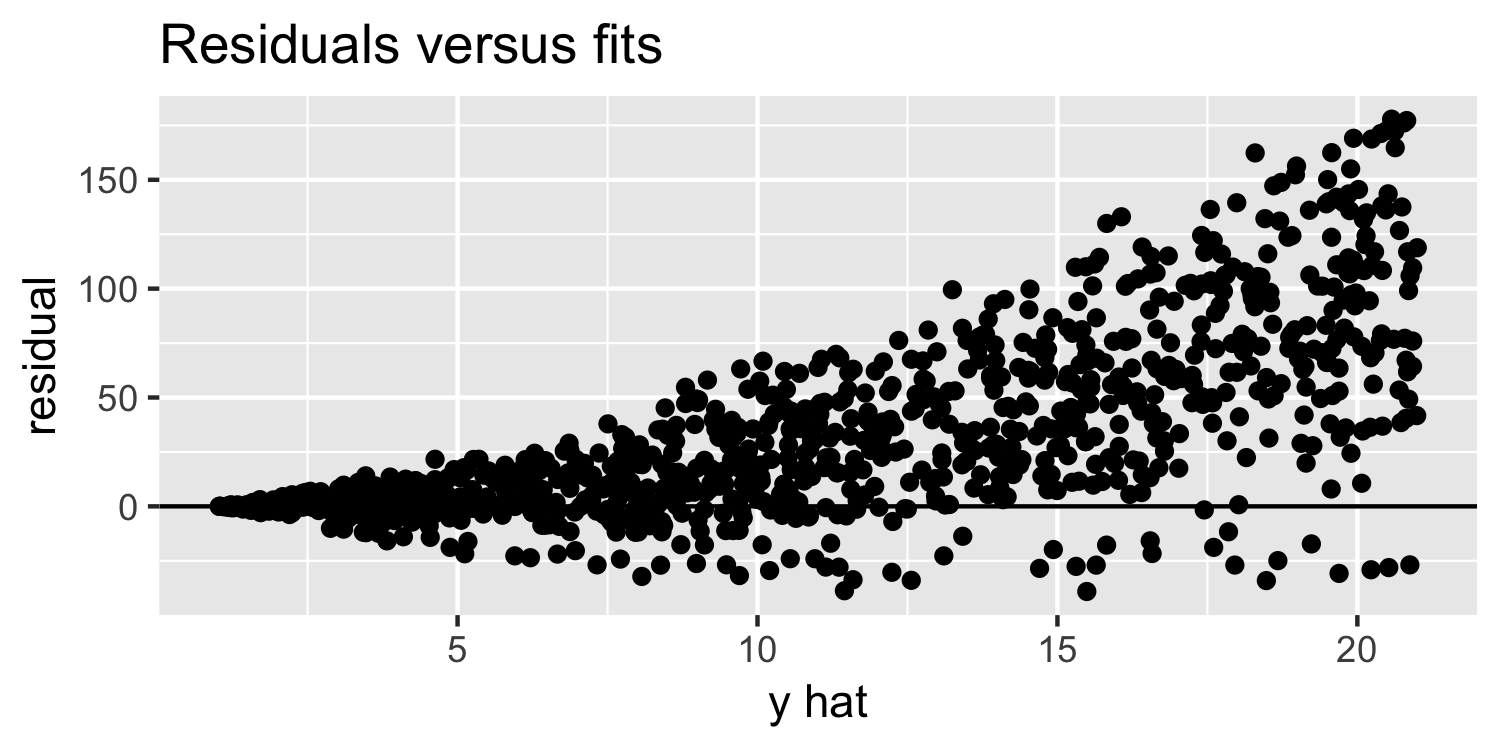
16 / 38
Dr. Lucy D'Agostino McGowan
Conditions for simple linear regression
- Linearity
- Zero Mean
- Constant Variance
- Independence
- Random
- Normality
17 / 38
Dr. Lucy D'Agostino McGowan
Conditions for simple linear regression
- Linearity
- Zero Mean
- Constant Variance
- Independence
- Random
- Normality
18 / 38
Dr. Lucy D'Agostino McGowan
Conditions for simple linear regression
- Linearity
- Zero Mean
- Constant Variance
- Independence
- Random
- Normality
19 / 38
Dr. Lucy D'Agostino McGowan
Normality
- Histogram
- Normal quantile plot
20 / 38
Dr. Lucy D'Agostino McGowan
Normality
Histogram
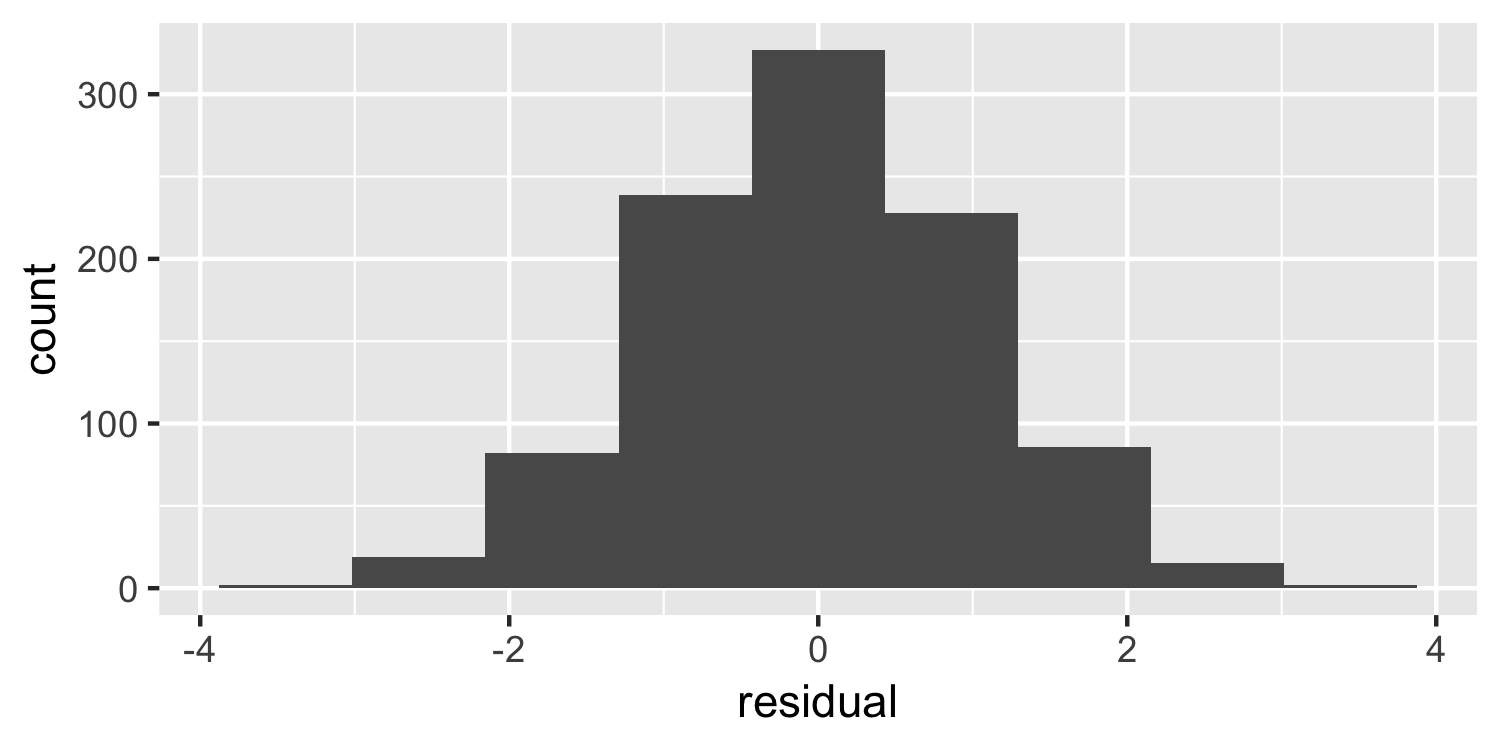
21 / 38
Dr. Lucy D'Agostino McGowan
Normality
Histogram
What do you think of this plot?
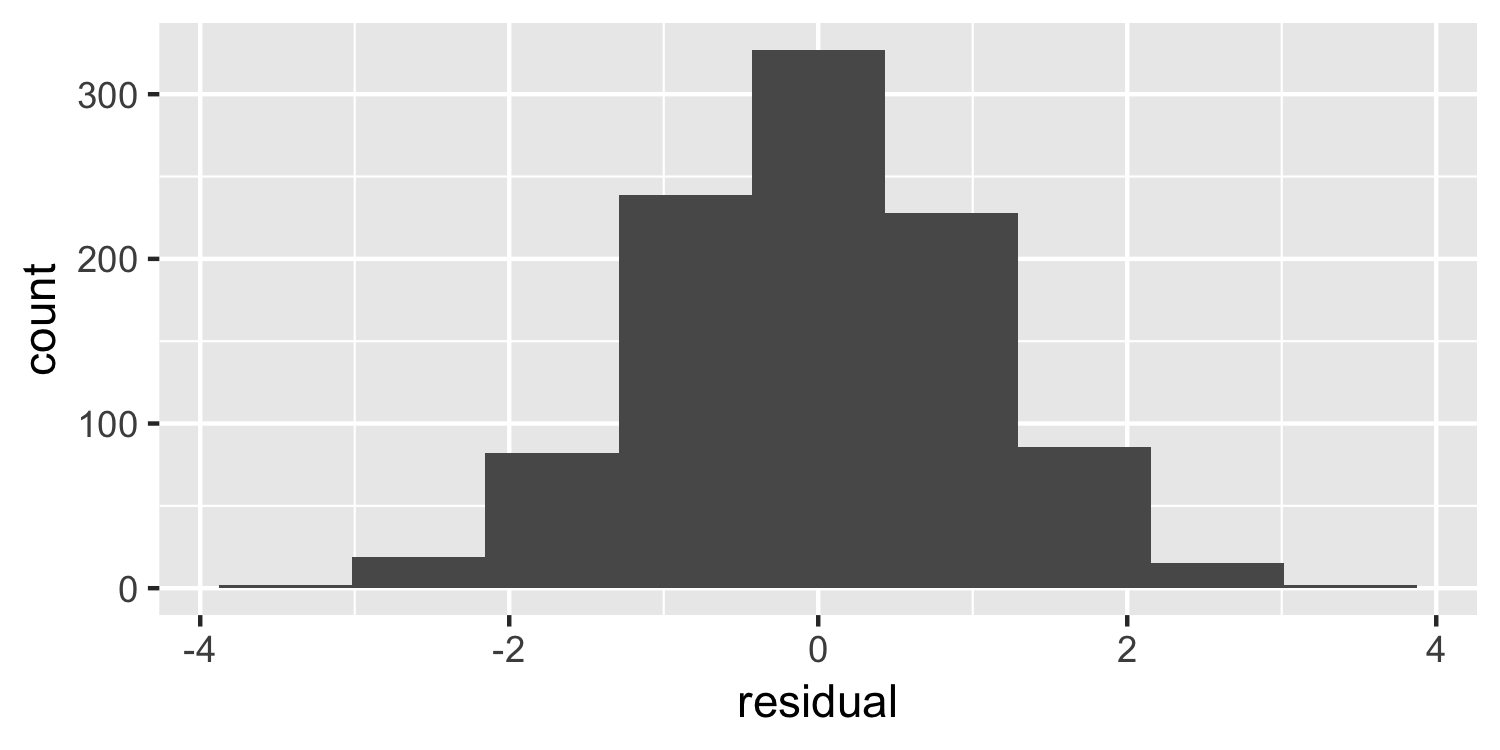
22 / 38
Dr. Lucy D'Agostino McGowan
Normality
Histogram
What do you think of this plot?
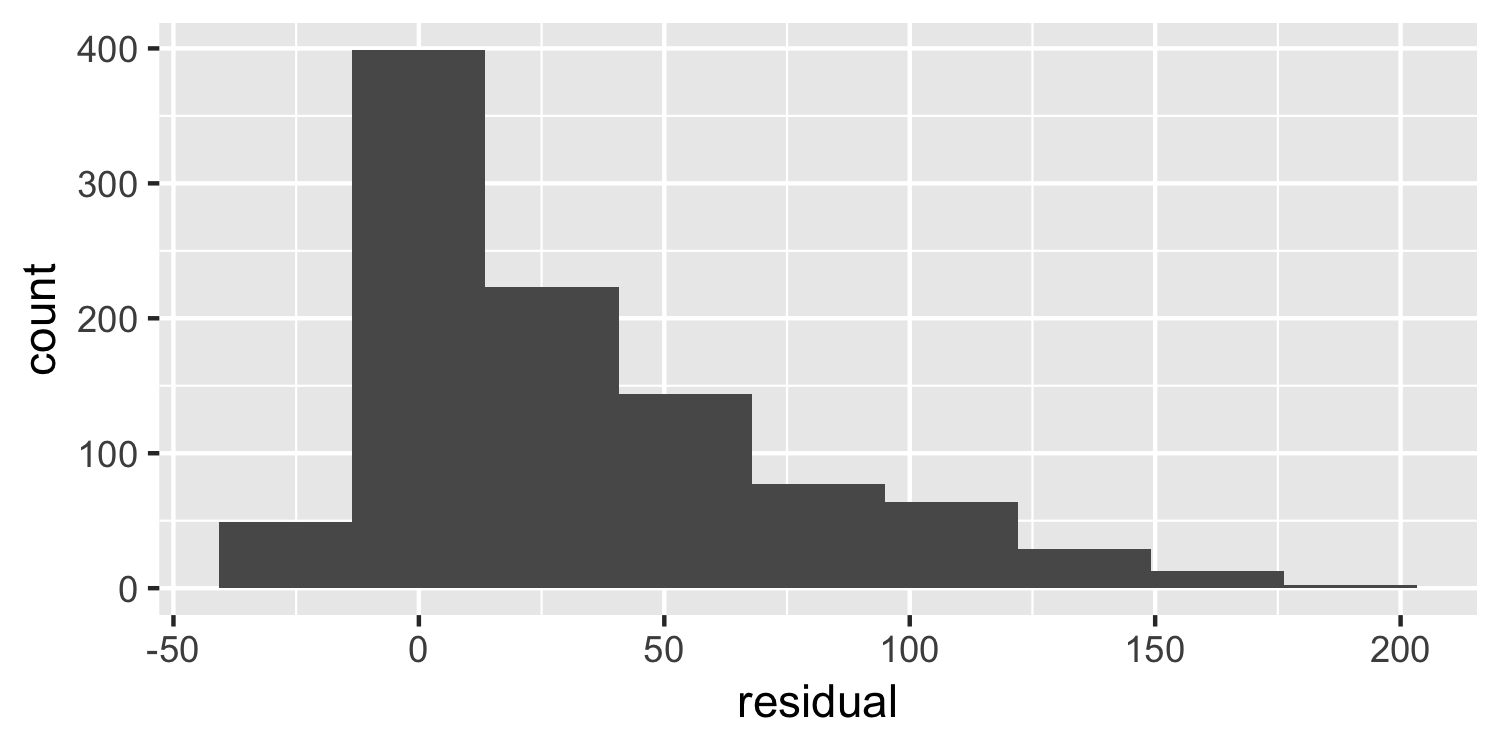
23 / 38
Dr. Lucy D'Agostino McGowan
Normality
Normal quantile plot
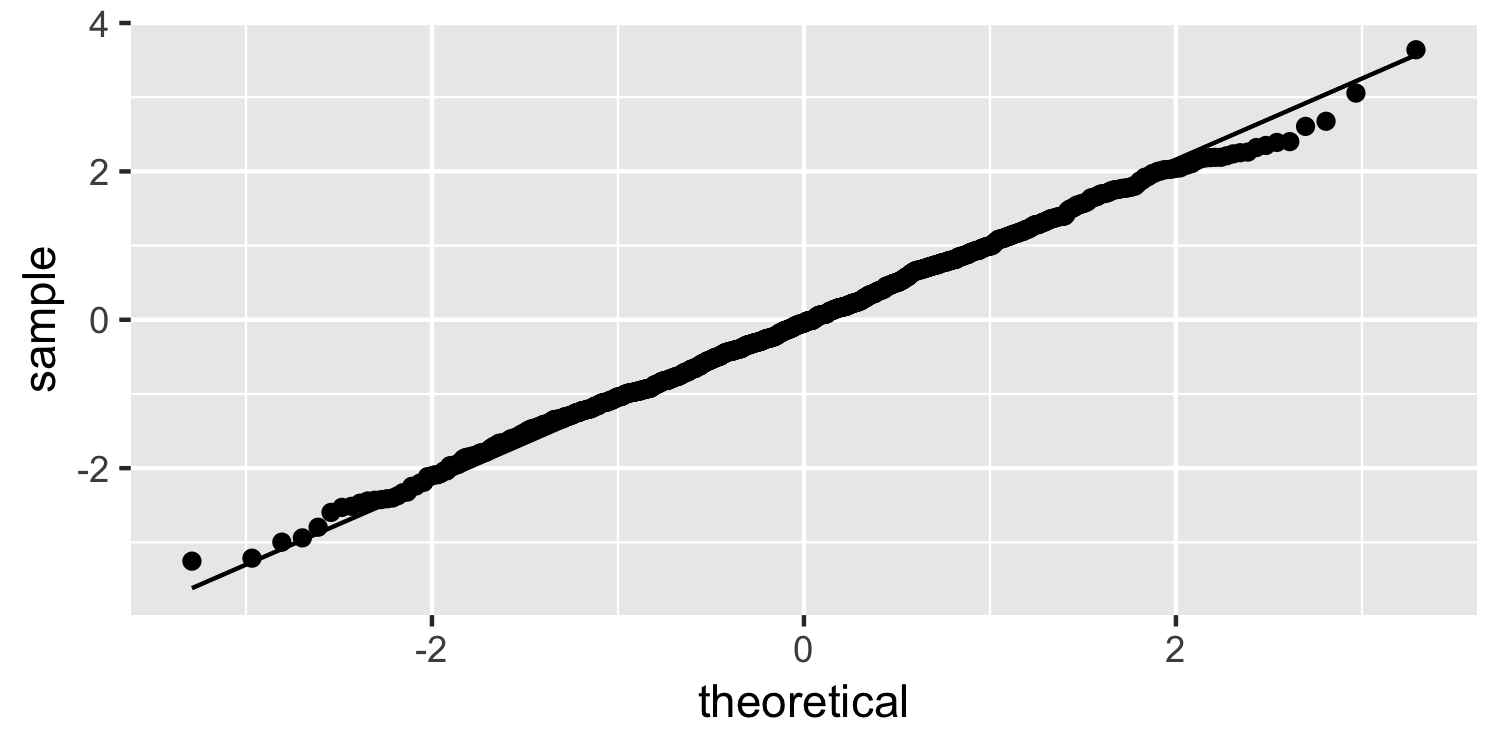
24 / 38
Dr. Lucy D'Agostino McGowan
Normality
Normal quantile plot
What do you think of this plot?
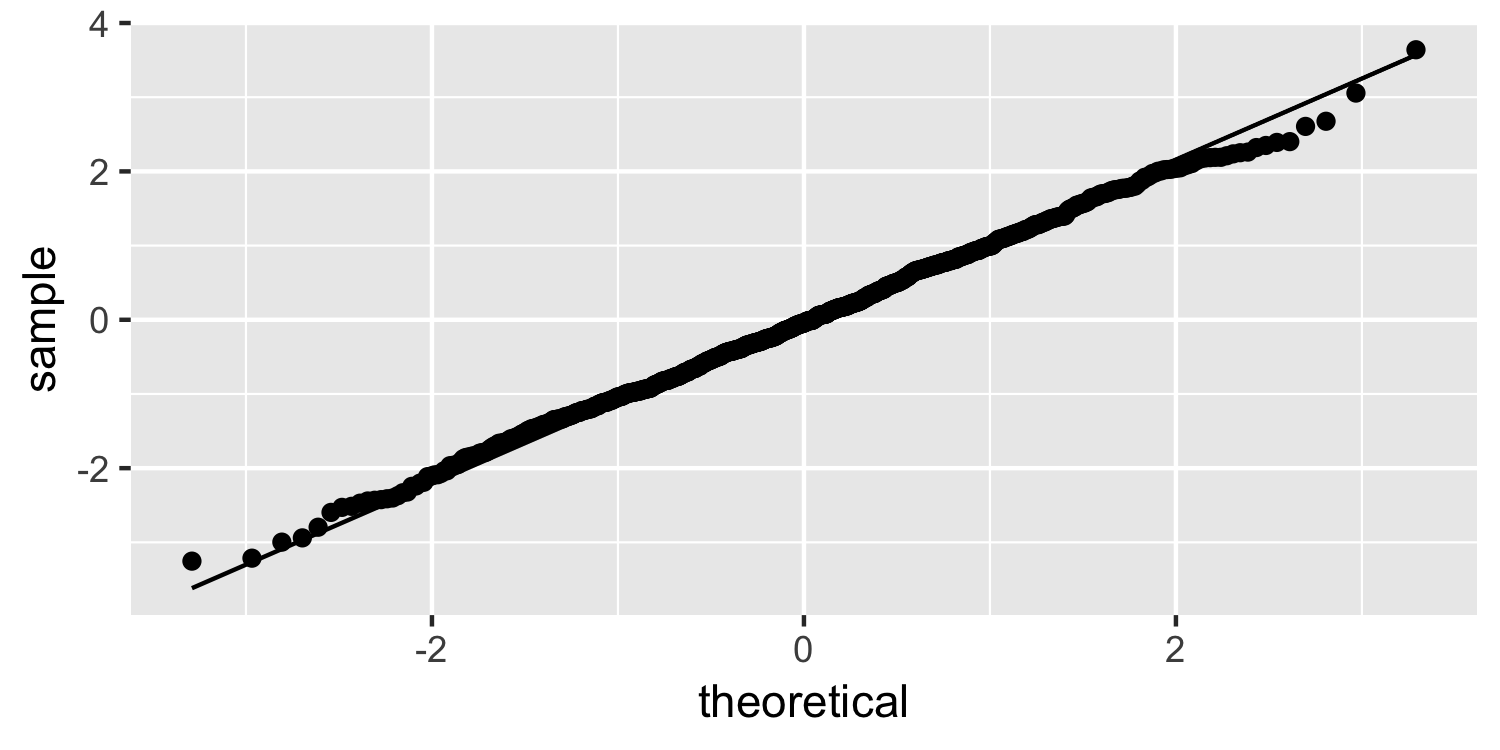
25 / 38
Dr. Lucy D'Agostino McGowan
Normality
Normal quantile plot
What do you think of this plot?
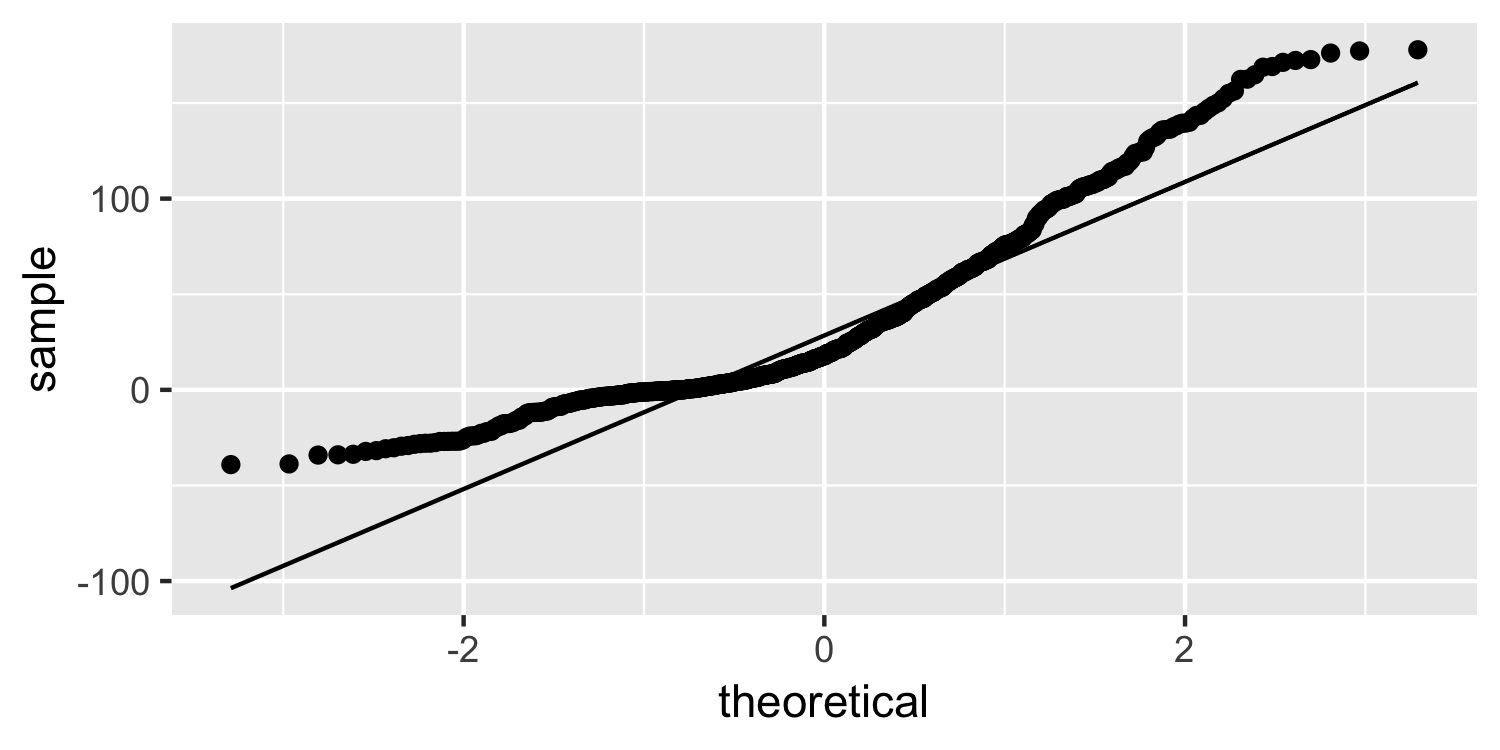
26 / 38
Dr. Lucy D'Agostino McGowan
Let's do it in R!
27 / 38
Dr. Lucy D'Agostino McGowan
Plot the data with a line
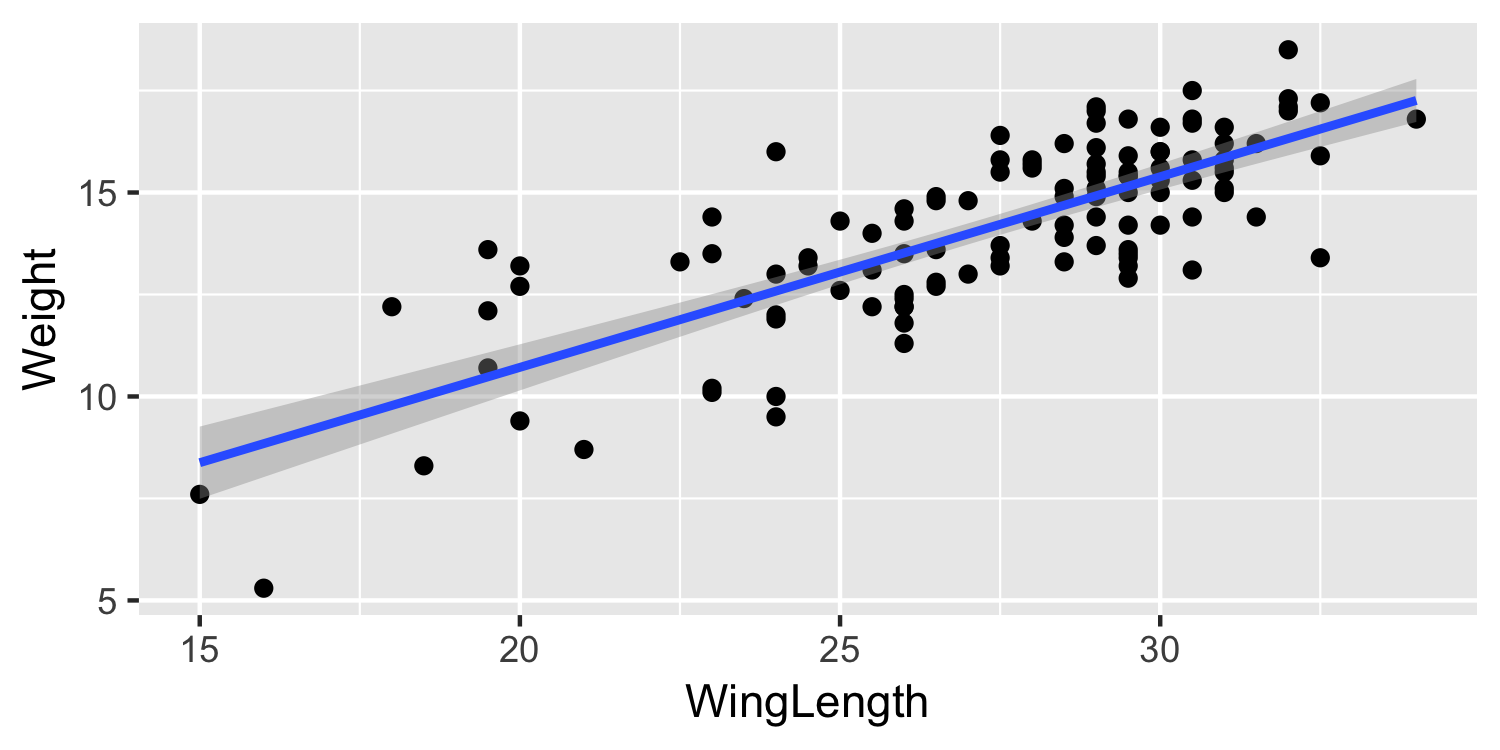
28 / 38
Dr. Lucy D'Agostino McGowan
Plot the data with a line
y_hat <- lm(Weight ~ WingLength, data = Sparrows) %>% predict()Sparrows <- Sparrows %>% mutate(y_hat = y_hat, residuals = Weight - y_hat)29 / 38
Dr. Lucy D'Agostino McGowan
Plot the data with a line
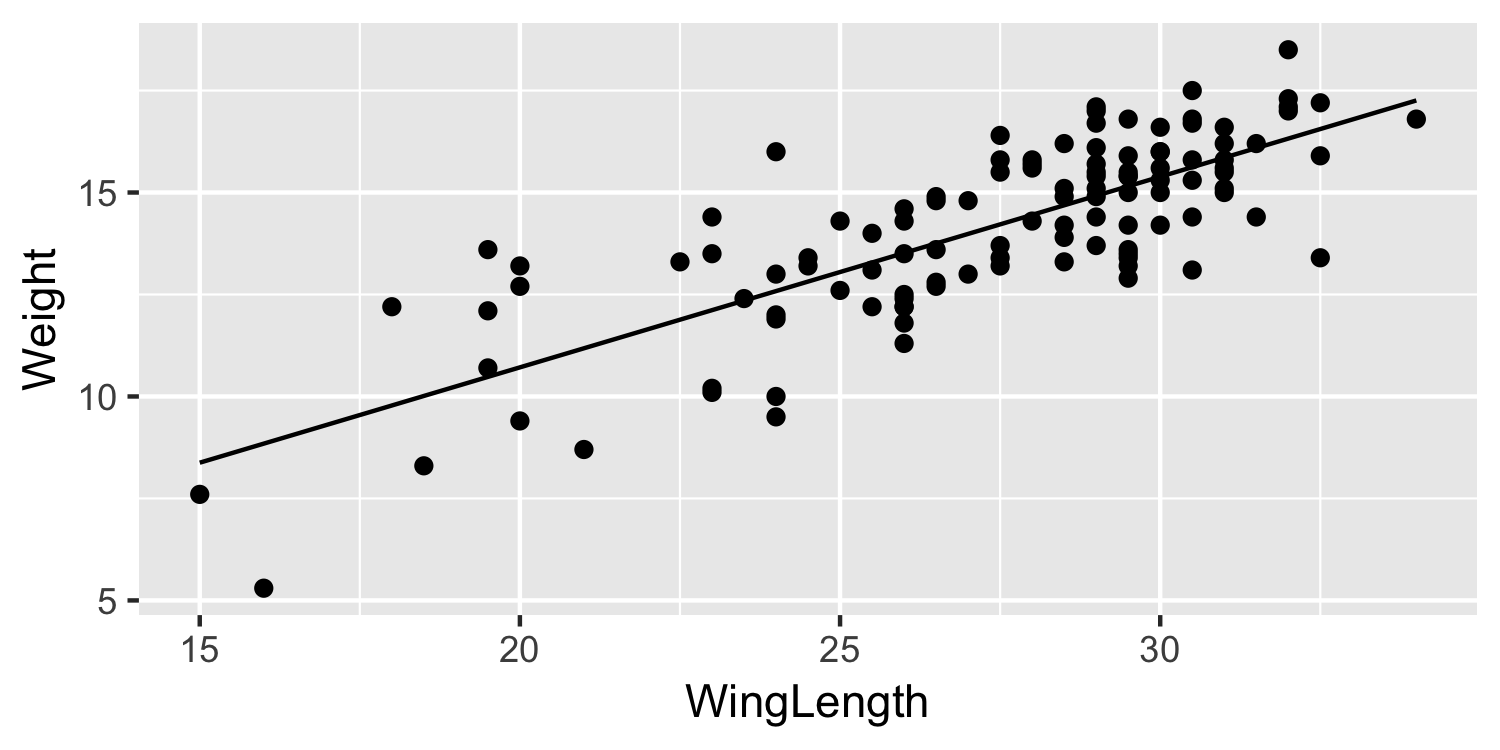
30 / 38
Dr. Lucy D'Agostino McGowan
Residuals vs Fits plot
ggplot(Sparrows, aes(x = y_hat, y = residuals)) + geom_point() + geom_hline(yintercept = 0) + labs(title = "Residuals versus fits", subtitle = "The relationship between WingLength and Weight for Sparrows", x = "y hat")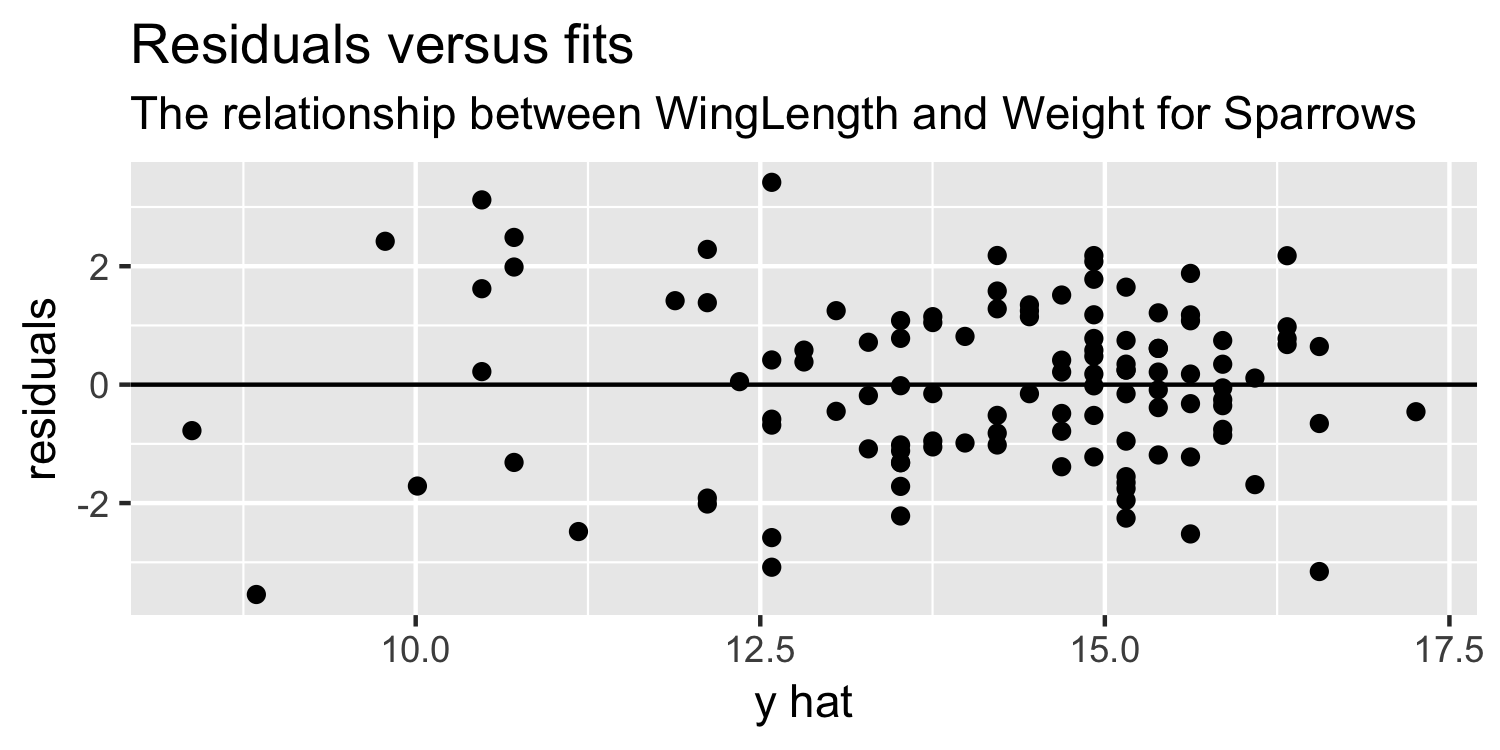
31 / 38
Dr. Lucy D'Agostino McGowan
Residuals vs Fits plot
ggplot(Sparrows, aes(x = y_hat, y = residuals)) + geom_point() + geom_hline(yintercept = 0) + labs(title = "Residuals versus fits", subtitle = "The relationship between WingLength and Weight for Sparrows", x = "y hat")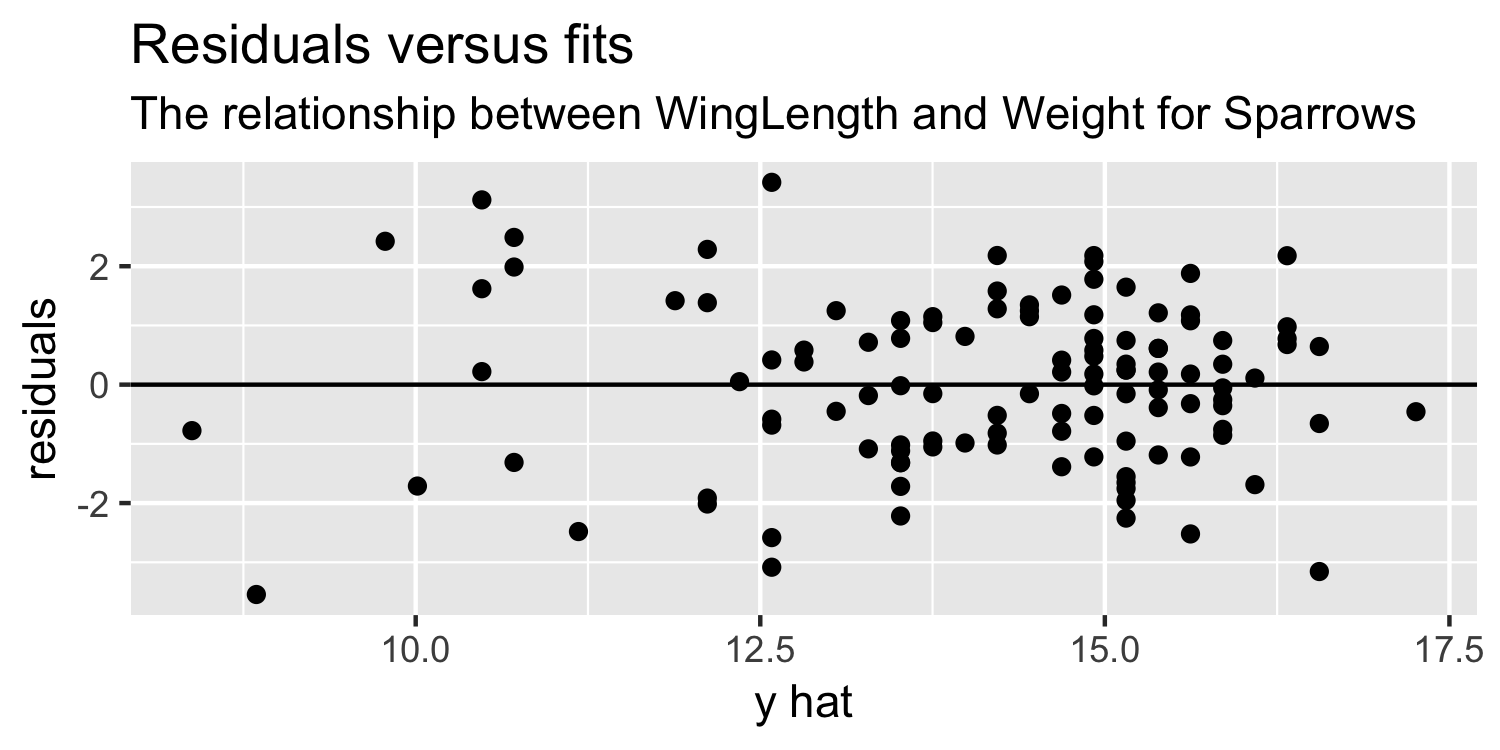
32 / 38
Dr. Lucy D'Agostino McGowan
Histogram of the residuals
ggplot(Sparrows, aes(residuals)) + geom_histogram(binwidth = 0.5)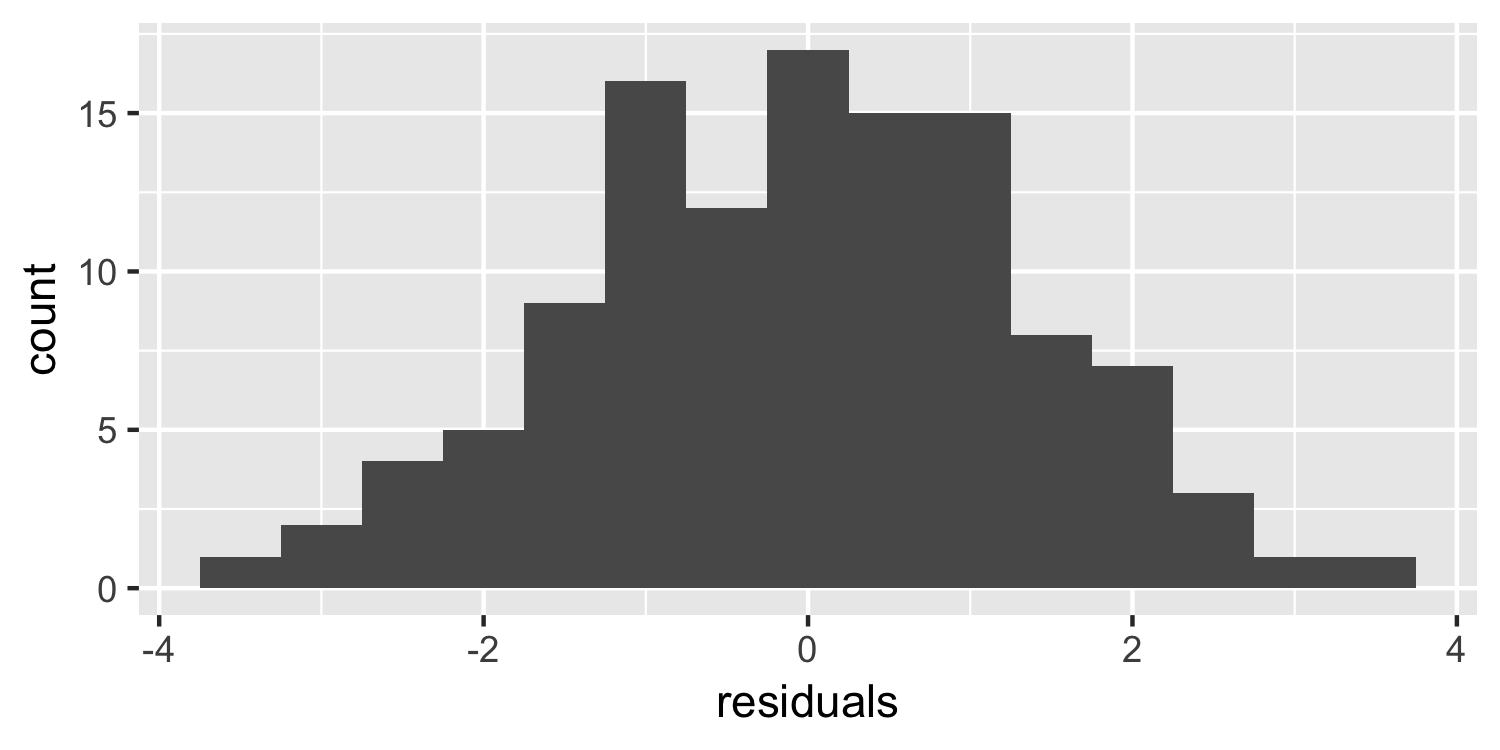
33 / 38
Dr. Lucy D'Agostino McGowan
Normal quantile plot
ggplot(Sparrows, aes(sample = residuals)) + geom_qq() + geom_qq_line()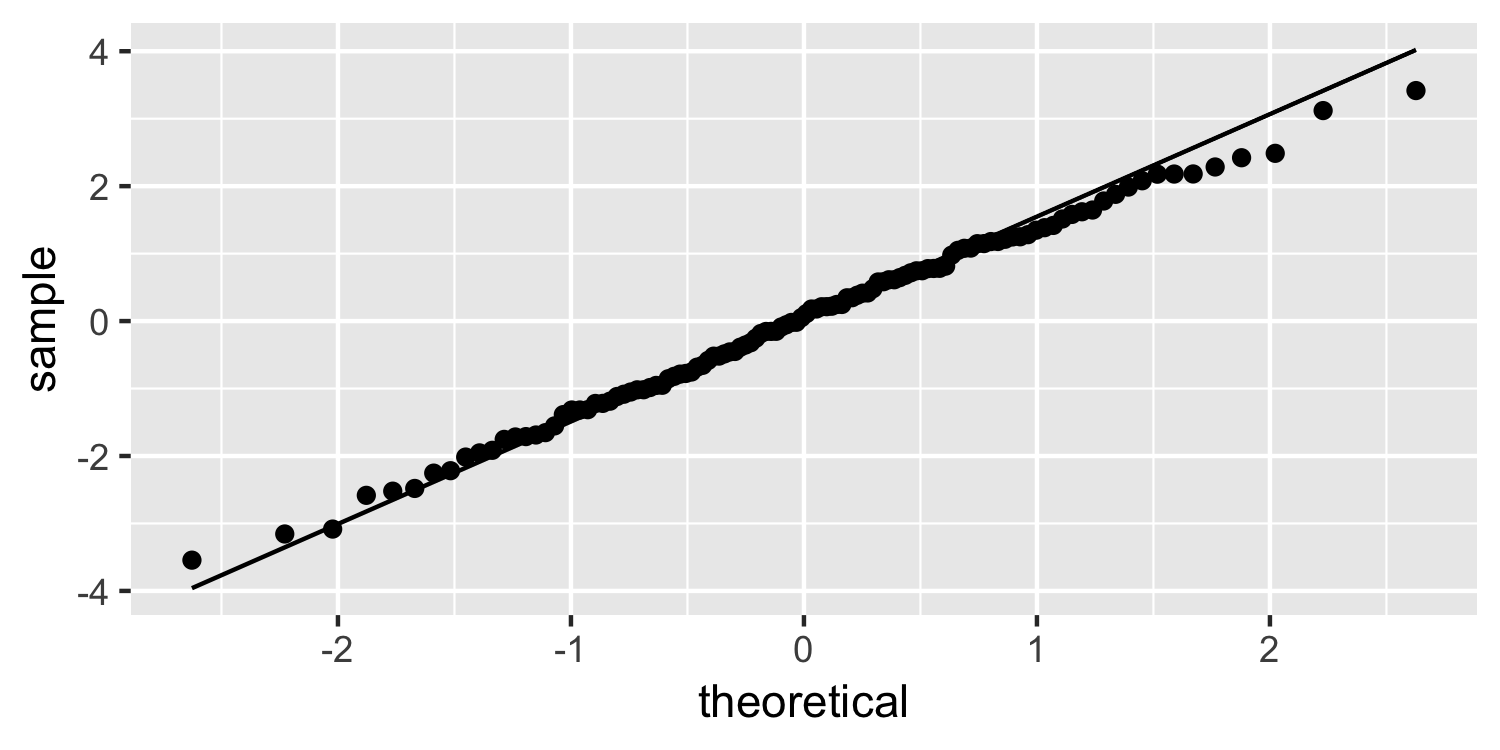
34 / 38
Dr. Lucy D'Agostino McGowan
Normal quantile plot
ggplot(Sparrows, aes(sample = residuals)) + geom_qq() + geom_qq_line()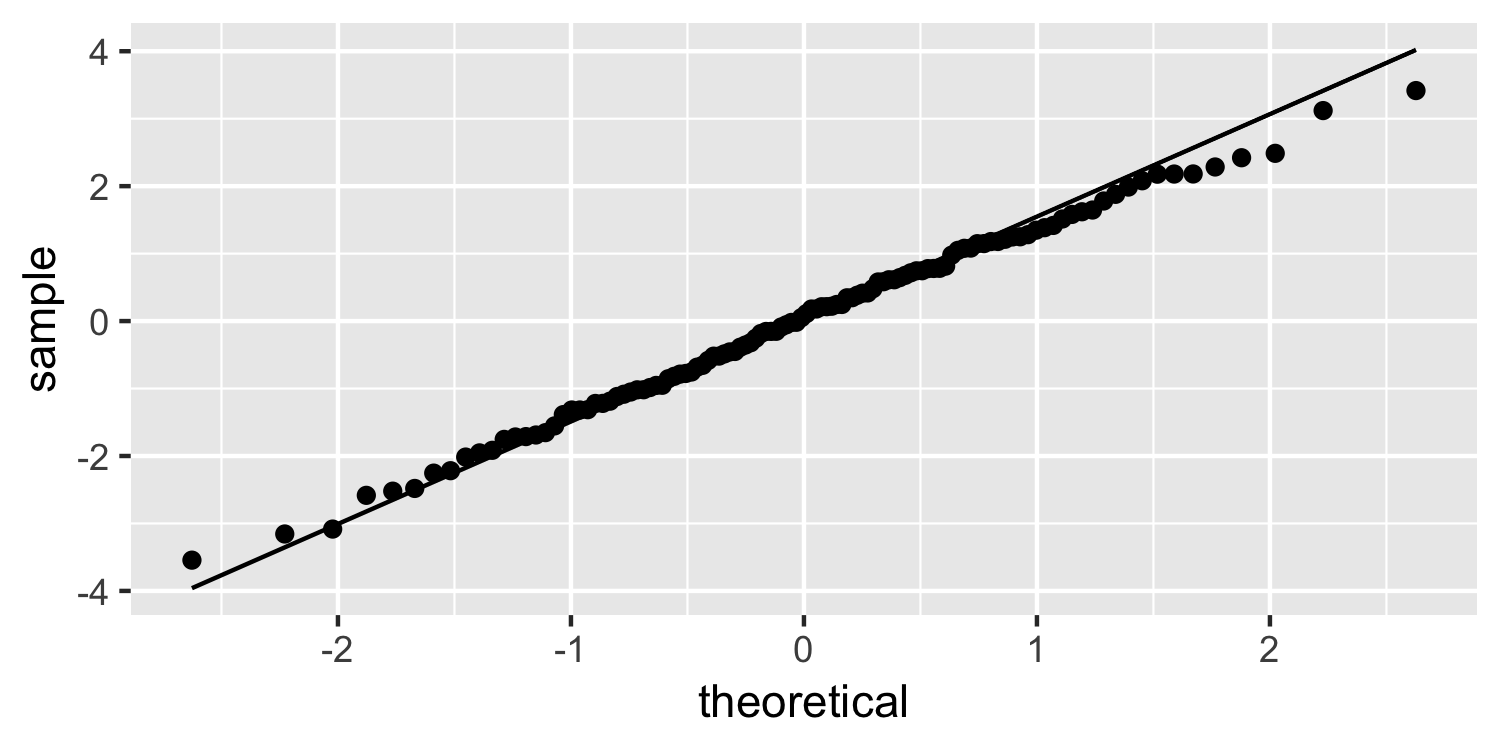
35 / 38
Dr. Lucy D'Agostino McGowan
Normal quantile plot
ggplot(Sparrows, aes(sample = residuals)) + geom_qq()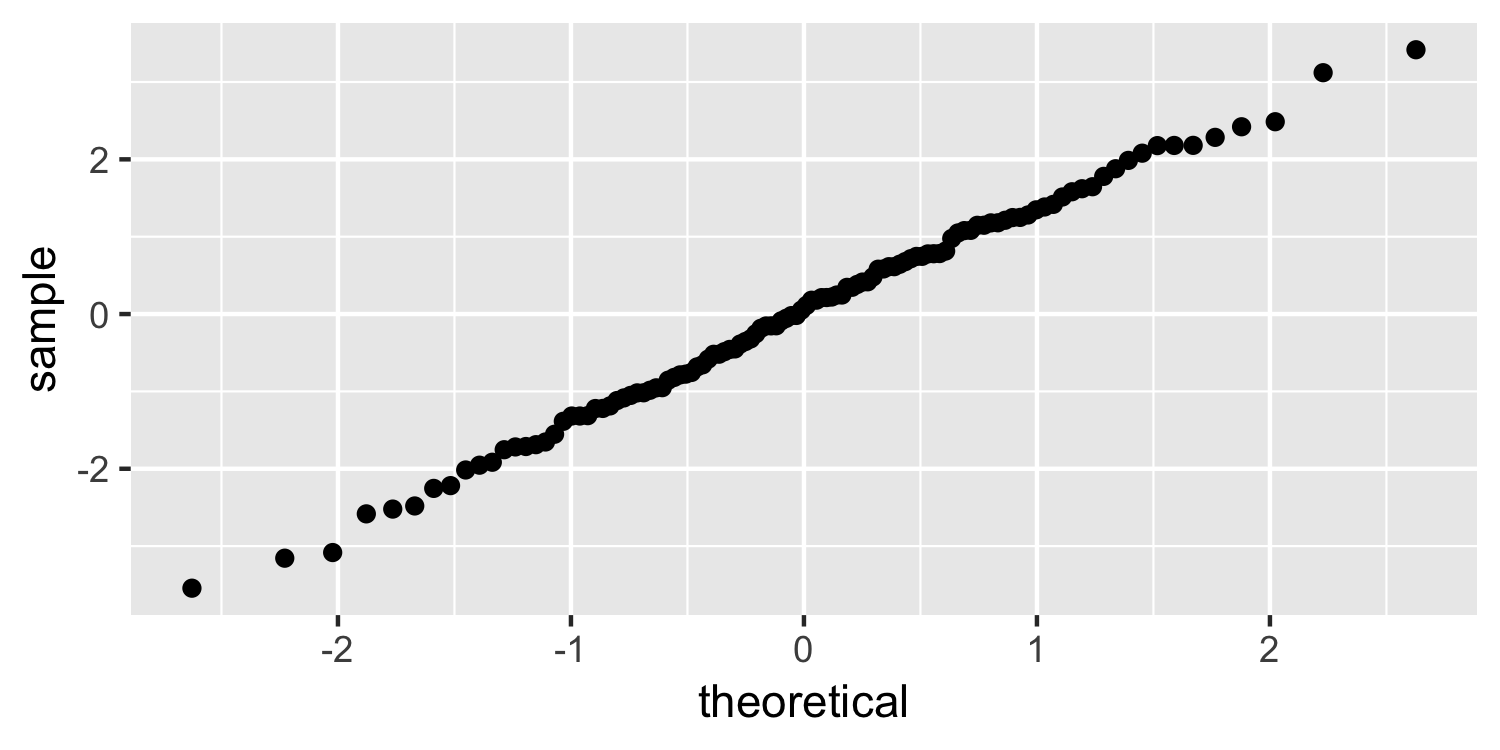
36 / 38
Dr. Lucy D'Agostino McGowan
Normal quantile plot
ggplot(Sparrows, aes(sample = residuals)) + geom_qq() + geom_qq_line()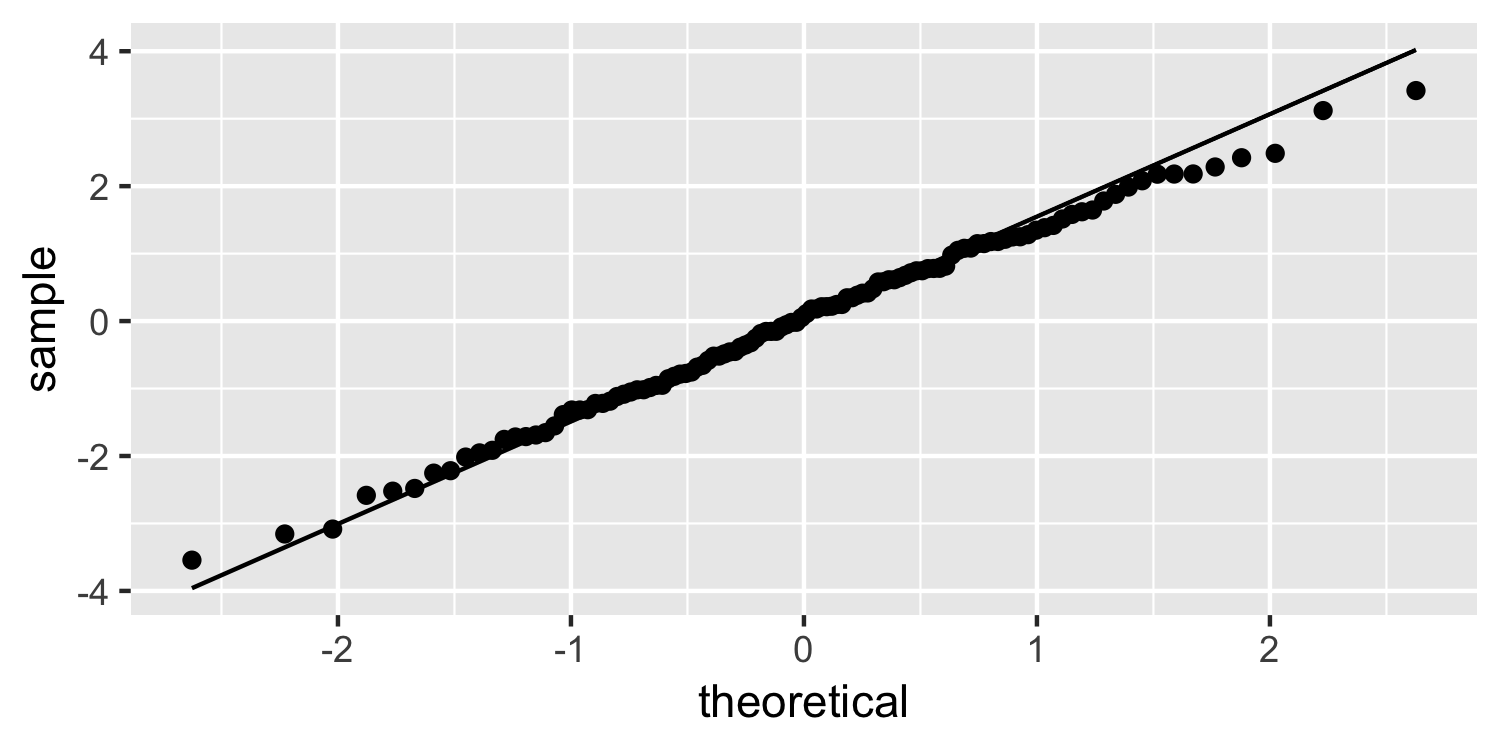
37 / 38
Dr. Lucy D'Agostino McGowan
Porsche Price (2)
- Go to RStudio Cloud and open
Porsche Price (2) - For each question you work on, set the
evalchunk option toTRUEand knit
38 / 38